Thermostable alkaline pectinase mutants and their coding genes and their applications
A technology encoding gene and pectinase, applied in the field of pectinase protein, can solve problems such as inability to withstand high temperature
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0034] Construction of pectinase mutant library by error-prone PCR method
[0035] Firstly, the PCR method was used (the upstream primer sequence of pelA is: 5'-GTGCGCTAGCATGGGACG-3', as shown in SEQ ID No: 9; the downstream primer sequence of pelA is 5'-GCGAAGCTTTCATCGATTTG-3', as shown in SEQ ID No: 10) from The pectinase Bsp165PelA gene pelA was amplified from the genome of alkalophilic Bacillus sp. N16-5 (CGMCC No.0369), and the recombinant plasmid pET28a-pelA was constructed. Using pET28a-pelA as a template, nucleotide mutations were introduced into pelA in vitro by using error-prone PCR technology. The sequences of random mutation primers are shown in SEQ ID No: 7 (MegaF: 5'-GCGGCAGCCATATGGCTAGCT-3) and SEQ ID No: 8 (MegaR: 5'-GCTCGAGTGCGGCCGCAAGCTTC-3).
[0036] The amplification system is:
[0037]
[0038] The PCR amplification conditions were: 94°C for 4min; 94°C for 45s, 55°C for 1min, 72°C for 1min, 30 cycles; 72°C for 10min.
[0039] The construction of the ...
Embodiment 2
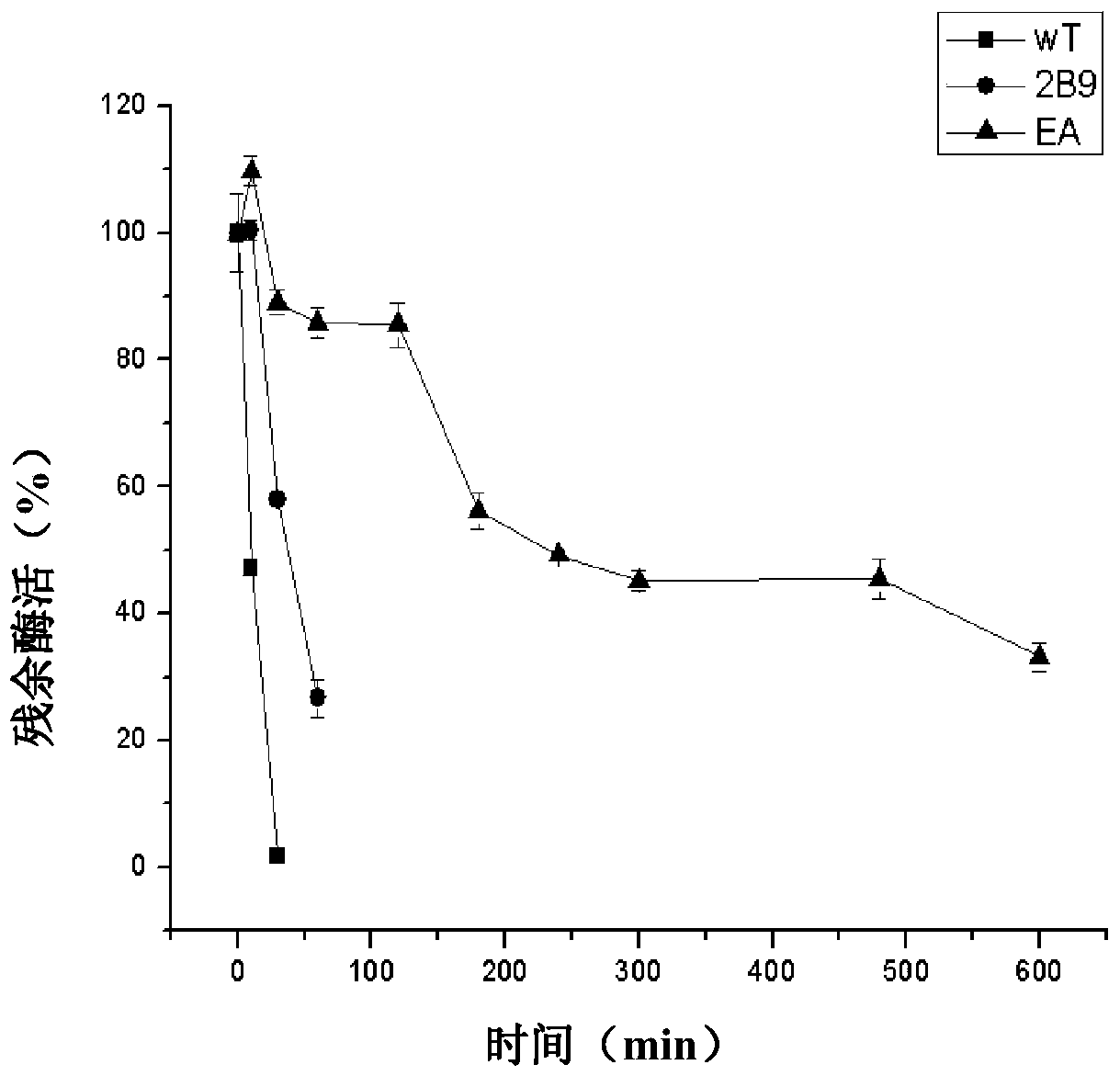
[0044] Screening of mutants with improved thermostability
[0045] The mutant and wild-type strains were replicated in fresh LB (containing 50ng / μL kanamycin, 0.1mM IPTG) liquid medium with a sterilized 96-well plate needle replicator, and induced overnight. The obtained bacterial fluid was used for further screening.
[0046] 1. Preliminary screening of 96-well plate: Dilute the cultured bacterial solution by 10 times, incubate at 65°C for 10 minutes, and then place it on ice to cool, absorb 20 μL of bacterial solution and 80 μL of 0.2% polygalacturonic acid (PGA) (w / v ) Glycin-NaOH buffer solution (pH10.0), mix well, react at 55 °C for 5 min, then cool on ice, add 80 μL DNS (3,5-dinitrosalicylic acid) solution, and react at 98 °C for 10 min, Determination of OD 540 . After being incubated at the selected temperature for 10 minutes, the residual enzyme activity of the wild type is basically 0, so the color is lighter; if the color of the mutant is darker after being incuba...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com