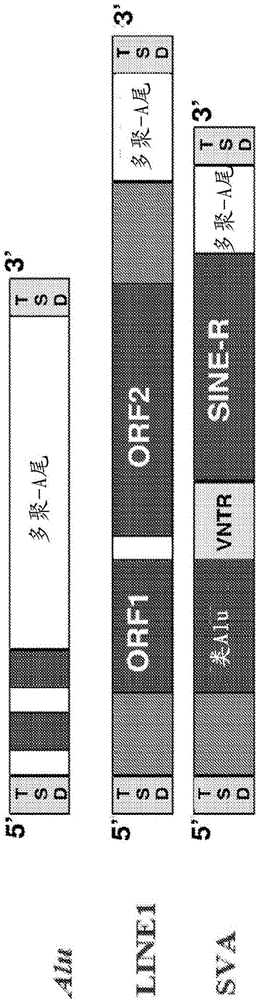
Method for genetic detection using interspersed genetic elements: multiplexed DNA analysis system
A technology for analysis and markers, which is applied in the field of human identification and biological ancestry testing, and can solve the problems of RE unavailability and size difference
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0081] Example 1: Four dye multiplex system for forensic applications.
[0082] A few markers are selected for multiplexing that can be used in forensic kits. Using three fluorophores, 6-carboxyfluorescein (6-FAM TM ), 4,5-dichloro-dimethoxy-fluorescein (JOE TM ) or carboxytetramethylrhodamine (TAMRA TM ) (using 5-carboxy-X-rhodamine (ROX TM ) and a fifth fluorophore at an orange wavelength as a size standard) to label the forward primer for each marker. The amplicon sizes of the selected markers ranged from about 46-124 bp, and the amplicon sizes of the individual INNUL alleles varied by 3-8 bp. The sex marker amelogenin was also added to the multiplex. Multiplex optimization experiments to address primer concentration and peak height issues were performed.
[0083] Markers were selected from http: / / dbRIP.org, existing literature, and by BLAST sequence analysis (A.F.A. Smit et al.; Batzer, M.A. et al. (2002); Batzer, M.A. et al. (1994); Feng, Q. et al.; Houck , C.M. et...
Embodiment 2
[0086] Example 2: Primer Design
[0087] Primers were designed using Primer3 (input version 0.4.0, http: / / frodo.wi.mit.edu / primer3 / ). Sets of three primers were designed for each marker: one forward primer and two reverse primers, one for the insertion allele and one for the null allele. All designed primers had Tm values in the range of 58-61°C. The program "Reverse Complement" from the Harvard Medical Technology Group and the Lipper Center for Computational Genomics (arep.med.harvard.edu / ) was used. Primers were then screened against GenBank's non-redundant database (National Center for Biotechnology Information, U.S. National Library of Medicine, National Institutes of Health) to determine if they were unique DNA sequences). Table 1 provides available markers and Table 2 provides primer sequences for selected markers.
[0088] Table 1. RE markers available for selection.
[0089]
[0090]
[0091]
[0092] Table 2. Primer sequences used for each INNUL marker ...
Embodiment 3
[0097] Example 3: Primer Preparation
[0098] Fluorescently labeled or unlabeled oligonucleotide primers were synthesized by Eurofins MWG Operon (Huntsville, AL, USA) or Integrated DNA Technologies (Skokie, IL). All lyophilized primers (labeled or unlabeled) were dissolved in 10 mM TE (tris(hydroxymethyl)aminomethane (“Tris”) and ethylenediaminetetraacetic acid (“EDTA”)) buffer (pH 8.0 ) to 100 μM stock solution concentration (10×). Store the stock primers at 4°C until use. After reconstitution, each primer was diluted to a final concentration of 10 μM (1×) using TE buffer. Each primer mix consisted of three primers: a labeled forward primer and two corresponding unlabeled reverse primers. The combined volume of the two reverse primers is equal to the volume of the forward primer. Store all labeled primers in opaque polypropylene tubes to avoid quenching of fluorescent labels.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com