Molecular biological method for rapidly detecting and identifying tomato spotted wilf virus and primer thereof
A technology of tomato spotted wilt virus and molecular biology, which is applied in the field of molecular biology detection and identification of plant viruses, can solve the problems of false positives, time-consuming, and labor-consuming, and achieve the elimination of false positives, wide detection range, and specificity strong effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0034] Example 1 The special primer L3 of the present invention for rapid detection and identification of tomato spotted wilt virus and its design and the molecular biology method for detecting and identifying tomato spotted wilt virus with the special primer L3
[0035] (1) Special primer L3 and its design and synthesis
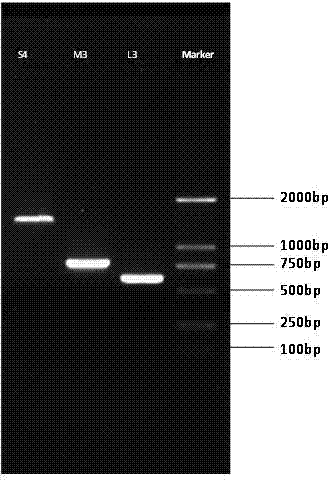
[0036] The special primer L3 is based on the RNA L fragment in the genome of Tomato spotted wilt virus (TSWV) (accession number: NC-002052) reported on NCBI (National Center for Biological Information, USA) as a template, and was designed using Primer 3 It is numbered as special primer L3, which is composed of a front primer and a back primer. The base sequence of the front primer of the special primer L3 is shown in SEQ ID NO: 1, and the back primer of the special primer L3 is The base sequence is shown in SEQ ID NO: 2, and the target product fragment length of the special primer L3 is 605bp.
[0037] The above special primer L3 was synthesized by Beijing ...
Embodiment 2
[0054] Example 2 The special primer M3 for fast and accurate detection and identification of tomato spotted wilt virus of the present invention and its design and molecular biology method for detecting and identifying tomato spotted wilt virus with the special primer M3
[0055] (1) Special primer M3 and its design and synthesis
[0056] The special primer M3 is based on the RNA M fragment in the genome of the tomato spotted wilt virus (Tomato spotted wilt virus, TSWV, accession number NC_002050) reported on NCBI as a template, using the primer designed by Primer 5, and it is numbered as the special primer M3, It is composed of a front primer and a back primer. The base sequence of the front primer of the special primer M3 is shown in SEQ ID NO: 3, and the base sequence of the back primer of the special primer M3 is shown in SEQ ID NO: 4. It is shown that the target product fragment length of the special primer M3 is 794bp.
[0057] The above special primer M3 was synthesized...
Embodiment 3
[0075] Example 3 The special primer S4 of the present invention for rapid and accurate detection and identification of tomato spotted wilt virus and its design and the molecular biology method for detecting and identifying tomato spotted wilt virus with the special primer S4
[0076] (1) Special primer S4 and its design and synthesis
[0077] Special primer S4 is based on the RNA S fragment in the genome of tomato spotted wilt virus (TSWV, accession number NC_002051) downloaded from NCBI as a template, and the primer designed by Primer5 is numbered as special primer S4, It is composed of a front primer and a back primer. The base sequence of the front primer of the special primer S4 is shown in SEQ ID NO: 5, and the base sequence of the back primer of the special primer S4 is shown in SEQ ID NO: 6. It is shown that the target product fragment length of the special primer S4 is 1277bp.
[0078] The special primer S4 mentioned above was synthesized by Beijing Sanbo Polygala Bio...
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com