Composition for estimating risk of onset of systemic lupus erythematosus, comprising primer for detecting dna copy number variation in 1q25.1(RABGAP1L) location, 6P21.32 (c4) location and 10.q21.3 location
A technology for copy number variation and lupus erythematosus, which is applied in the direction of recombinant DNA technology, DNA/RNA fragments, genetic material components, etc., to prevent the deterioration of the state and reduce the effect of analysis errors
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0042] Embodiment 1. Research object and statistical analysis method
[0043] 1-1. Research object
[0044] All subjects were composed of Korean ethnicity.
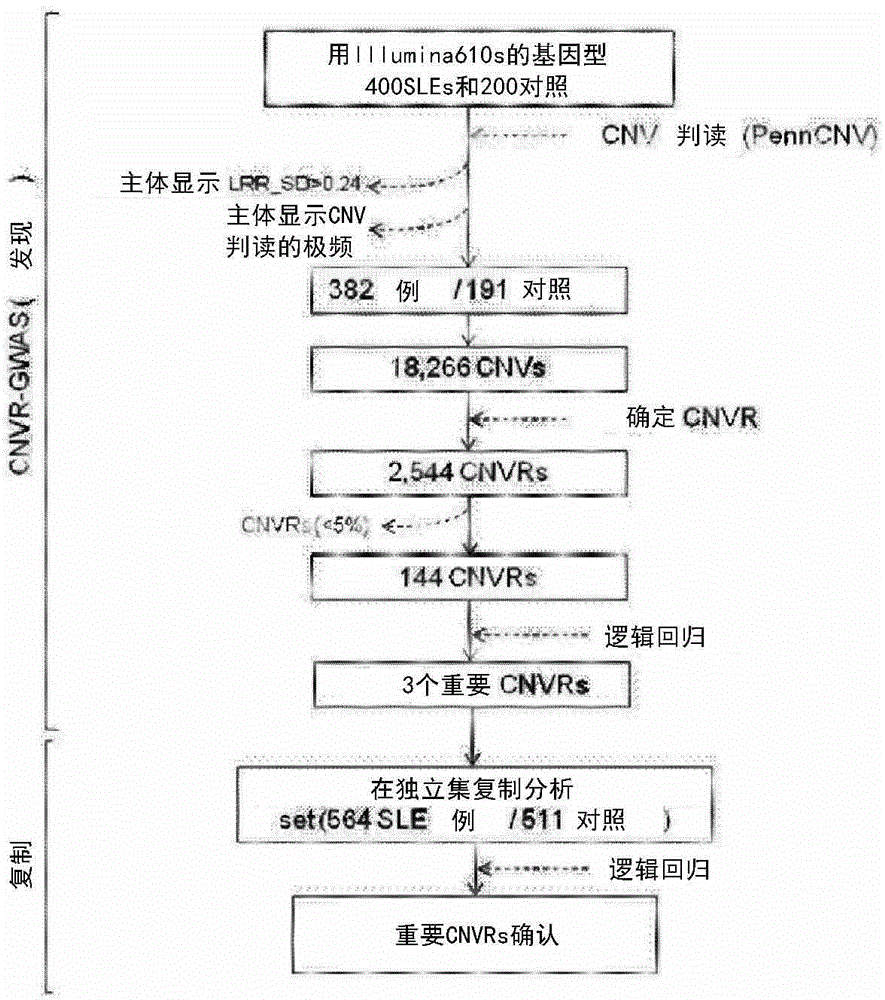
[0045] For CNV-GWAS analysis, 400 SLE patients (all female, mean age 31±10.0 years) were recruited from the BAE systemic lupus erythematosus population of the Department of Rheumatic Diseases, Hanyang University Hospital (Seoul, Republic of Korea). These patients were all patients diagnosed as suffering from SLE through the 1997 American College of Rheumatology classification criteria (American College of Rheumatology classification criteria). Meanwhile, 200 non-SLE patients were recruited from the same hospital as a control group (all female, mean age 33±9.2 years).
[0046] For independent copies, 564 patients and 511 controls (1075 in total) were recruited from the same hospital.
[0047] This study was conducted after approval by the institutional review board (CUMC11U199).
[0048] 1-2. Statistical analysis...
Embodiment 2
[0050] Example 2. Whole genome SNP genotype (genotype) analysis
[0051] The human 610s Quad-Bead Chip platform (including 620,901 SNP markers) produced by Illumina was used to perform genotype analysis on the whole genome SNP. For each known CNV, the average number of probes was 37.7. Approximately 750 ng of genomic DNA per sample was used for genotype analysis. The samples used by the inventors showed an average SNP call rate of 99.89±0.16%. All steps were performed in the same environment at the same time in order to minimize plate or batch effects.
Embodiment 3
[0052] Example 3. Quality verification and copy number variation (CNV) exploration
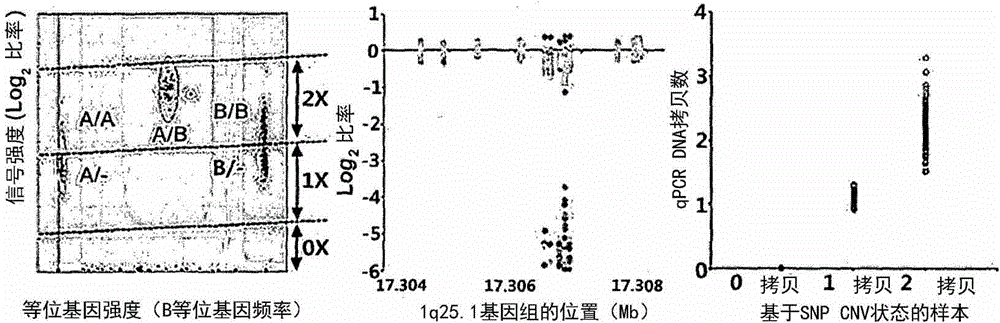
[0053] The experimental data including signal intensity (log R ratio: LRR) and allele intensity (B allele frequency) were obtained using GenomeStudio software produced by Illumina.
[0054] 3-1. Quality Verification
[0055] First, a process of verifying the quality of said data was passed. In the following cases, the relevant individuals were excluded from the experimental results: when the successful SNP sampling rate (SNP interpretation rate) was less than 90% or the LRR standard deviation exceeded 0.24; when the B zygote frequency drift (drift) exceeded 0.01; When the wave factor (wave factor) is less than 0.05.
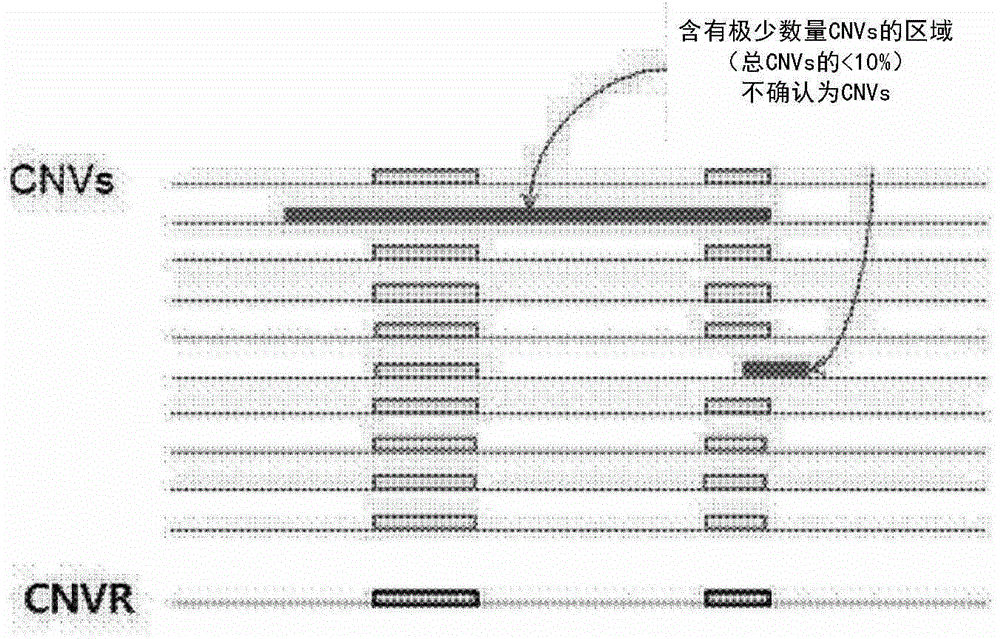
[0056] 3-2. CNV sampling
[0057] The present inventors sampled CNV based on signal intensity (log R ratio: LRR) and allele intensity (B allele frequency) data and using the PennCNV algorithm as the basic setting state.
[0058] The PennCNV algorithm uses HMM, although ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com