siRNA for inhibiting ADAM17 (a disintegrin and metalloprotease 17) genes and application of siRNA
A DNA molecule, methoxyl modification technology, applied in DNA/RNA fragments, recombinant DNA technology, medical preparations containing active ingredients, etc., can solve problems such as differences in silencing effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
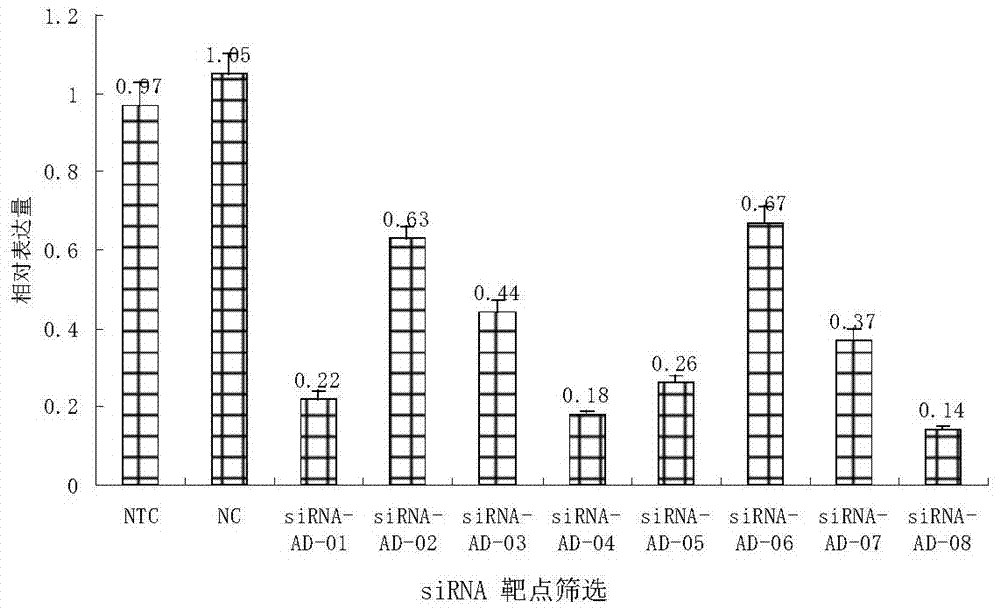
[0090] Embodiment 1, the screening of the effective oligonucleotide that suppresses ADAM17 gene mRNA expression
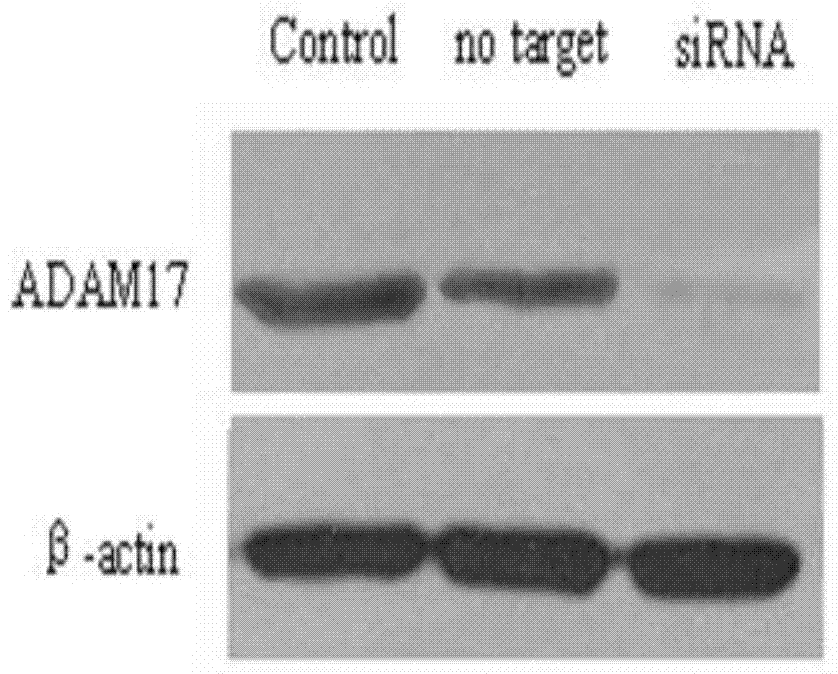
[0091] 1. Perform siRNA design to determine siRNAs targeting ADAM17, and perform bioinformatic screening to ensure that the sequence is specific to the ADAM17 sequence and not specific to sequences from any other gene. The target sequence was checked against the sequence in GenBank using the BLAST search engine provided by NCBI. After preliminary experiments, 8 effective siRNAs were screened out, named siRNA-AD-01, siRNA-AD-02, siRNA-AD-03, and siRNA respectively. - AD-04, siRNA-AD-05, siRNA-AD-06, siRNA-AD-07, siRNA-AD-08. The above siRNAs are siRNAs designed for different positions of the ADAM17 gene sequence.
[0092] 2. Cell transfection
[0093] The experiment was divided into 10 groups, namely siRNA-AD-01 to siRNA-AD-08 experimental groups, Notarget (NTC) was the negative control group, and NC was the blank control group.
[0094] The setting method of siR...
Embodiment 2
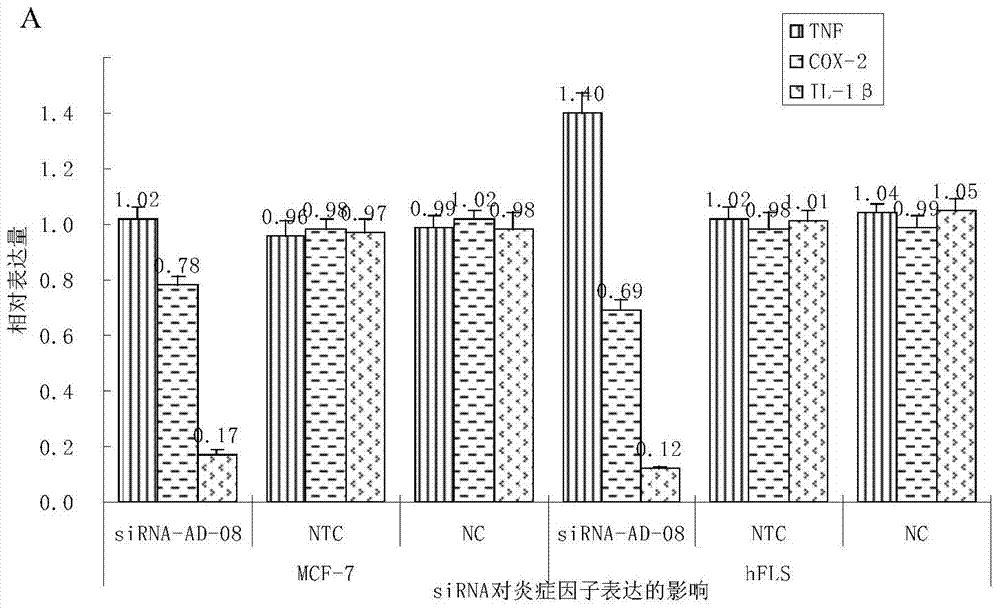
[0113] Embodiment 2, the inhibition of oligomeric nucleic acid to inflammatory factor
[0114] 1. The experiment is divided into the following groups:
[0115] hFLS-siRNA-AD-08 experimental group: primary culture hFLS cells to 6-well plate, when the cell density was about 50%, according to Lipofectamine2000 kit instructions, transfect hFLS cells with siRNA-AD-08 at a final concentration of 50nM.
[0116] MCF-7-siRNA-AD-08 experimental group: primary culture MCF-7 cells to 6-well plate, when the cell density is about 50%, according to Lipofectamine2000 kit instructions, transfer siRNA-AD-08 according to the final concentration of 50nM Infect MCF-7 cells.
[0117] hFLS-No target (NTC) negative control group: replace the siRNA in the hFLS-siRNA-AD-08 experimental group with random non-specific siRNA, and the rest of the steps remain unchanged. Among them, the random non-specific siRNA is not specific to the siRNA designed for the target gene (ADAM17 gene):
[0118] Sense stran...
Embodiment 3
[0144] Example 3, verification of homologous oligonucleotides on ADAM17 gene inhibitory effect
[0145] In order to verify the influence of the homology ratio on the effect of siRNA-AD-08 on suppressing the ADAM17 gene, the following three sets of experiments were carried out:
[0146] 1. The first set of experiments
[0147] The siRNA antisense strands of the first group are all "5'-UUGUUCAGAUACAUGAUGC-3'"
[0148] , the sense strand is the homologous sequence of "5'-GCAUCAUGUAUCUGAACAA-3'", as shown in Table 1.
[0149] Table 1 Antisense strand group
[0150]
[0151] Note: S=sense strand, AS=antisense strand. Choose 11nt, 15nt, 23nt, 27nt, and mismatches for the sense strands, respectively.
[0152] According to the method of Example 1, each siRNA shown in Table 1 was transfected into hFLS cells, and its inhibitory efficiency on ADAM17 gene mRNA expression was detected.
[0153] 2. The second set of experiments
[0154] The sense strands of the second group of siRN...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com