A Molecular Approach to Differentiate Cherry Fruit Color
A technology for cherries and fruits, which is applied in the molecular field of distinguishing the color of cherries, achieves the effects of simple operation, easy popularization, and beneficial to auxiliary breeding.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
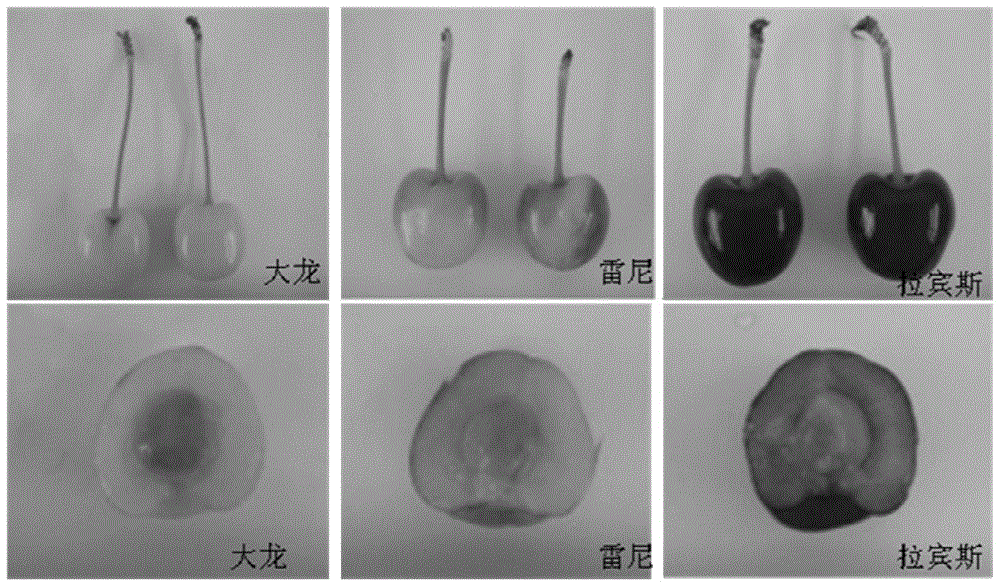
[0028] Example 1 Detection of the colors of Dalong, Rainey and Rabbins by the method of the present invention
[0029] The Dalong, Rainey and Rabbins leaves were selected as experimental materials. The color of the mature peel of Dalong is yellow and the flesh is yellow; the color of the mature peel of Raney is red with a yellow background and the flesh is light yellow; figure 1 ). The specific operations are as follows:
[0030] Use Biomiga's Plant gDNA kit to extract the genomic DNA of the three materials of cherry Dalong, Raney and Labins. The method refers to the kit, electrophoresis on 0.8% agarose gel (EB), and the extracted genomic DNA is observed under ultraviolet light ( See figure 2 ).
[0031] The gene LGS was amplified by PCR method, and the PCR reaction system was used to amplify the three materials of Dalong, Raney and Labins respectively.
[0032] The upstream primer is ATGGAGGGTATAACTTGGGTGTGAGAA (Sequence Listing NO.1);
[0033] The downstream primer i...
Embodiment 2
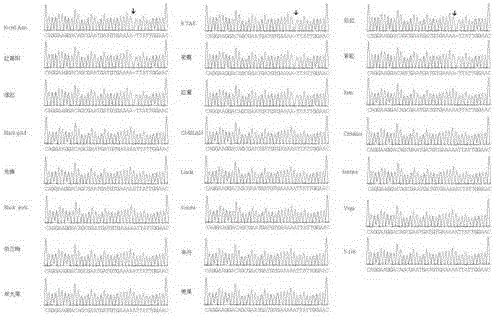
[0038] Embodiment 2 uses the method of the present invention to detect 26 cherries of different colors
[0039] Keeping other conditions unchanged, blind testing was carried out on 26 cherries of different colors, that is, the fruit color was inferred from the molecular test results, and the inferred results were confirmed by the color of the ripe fruit. The specific operations are as follows:
[0040] Use Biomiga's Plant gDNA kit to extract cherries Royal Ann, Chelan, Critalina, Sam, B.TAR, Jiahong, Hongnanyang, Caixia, Pioneer, Hongmi, Linda, Sonata, BlackYork, Juhong, Berrant, Santina, Zao Daguo, Zaodan, Vega, Black Gold, S-106, Rainbow, 133, Dalong, Raney and Rabins. The genomic DNA of 26 leaf materials, method reference kit.
[0041] The gene LGS was amplified by PCR method. PCR reaction system: 26 genomic DNAs were taken as templates for amplification.
[0042] The upstream primer is ATGGAGGGTATAACTTGGGTGTGAGAA;
[0043] The downstream primer is TAGTCCTTCTGAACATTGGTAC...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com