A systematic detection method of dna transcription direction and transcription template and its application
A detection method and technology of transcription direction, applied in the field of systematic detection of DNA transcription direction and transcription template, can solve the problems of cumbersome steps in northern blot, high cost and high cost of single-stranded probes, and achieve low price, low cost and fast speed Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
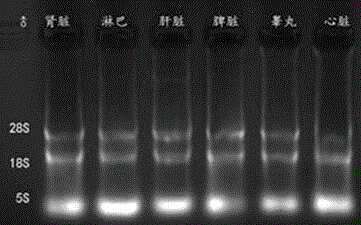
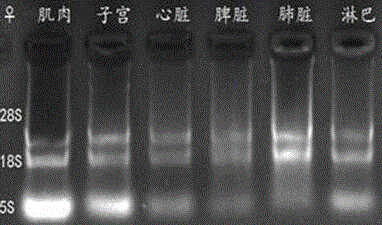
[0063] Example 1 Analysis of mouse β-actin and Tsix gene transcription direction
[0064] 1. Test materials and test methods
[0065] (1) Test animals
[0066] Kunming white mice, female mice 7-8 weeks old, male mice 3-6 months old, feeding conditions: light 6:00AM-8:00PM, temperature 20-25℃, free to eat.
[0067] (2) Main equipment and instruments
[0068] Scissors, tweezers, and absorbent cotton should be treated with RNase inhibitors before use; pipette tips, centrifuge tubes, and PCR tubes should all be treated with DECP water, and the concentration of DECP should be 0.1%. Ice maker, -20℃ refrigerator, -80℃ refrigerator, desktop centrifuge, high-speed refrigerated centrifuge, electrophoresis instrument, electronic analytical balance, PCR instrument.
[0069] (3) Experimental reagents
[0070] Mix (Transgene), markerD2000 (Genestar), reverse transcription kit (Promega), RNase inhibitor (Thermo), S1 nuclease (TaKaRa), Trizol (TaKaRa), DNase I (Thermo).
[0071] 50×TAE...
Embodiment 2
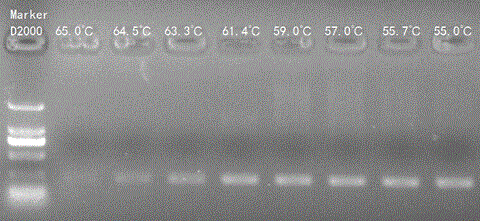
[0139] The total RNA in the chicken liver tissue was extracted and reverse-transcribed, and the method was the same as in Example 1. Use the software VectorNTI and primerpremier5 to design the primers GHR (LRP1)-F3 (sequence shown in SEQ ID NO.5) and GHR (LRP1)-R3 (sequence shown in SEQ ID NO.6) of the target gene GHR, and determine the suitable annealing temperature of the primers and reaction conditions. The primer information is shown in Table 8:
[0140] Table 8 Chicken GHR primer information
[0141]
[0142] GHR gene PCR reaction conditions are as follows:
[0143] 95°C, 5min→(94°C, 30sec→60°C, 30sec→72°C, 30sec) 40cycles→72°C, 5min→16°C, 1h.
[0144] Subsequent detection methods and operations are the same as in Example 1.
[0145] Experimental results such as Figure 11~13 shown. According to our experimental results, it is determined that the detected DNA segment is bidirectionally transcribed, and both strands of DNA can be used as templates to transcribe RN...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com