Application method of long-chain non-coding RNA CASC2 derived from serum exosomes
A long-chain non-coding, application method technology, applied in the application field of LncRNACASC2 in the diagnosis of glioma, can solve the problems of unsatisfactory early diagnosis and standardized treatment
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0023] Example 1 Preparation of long-chain non-coding RNACASC2 kit for screening, early diagnosis or prognosis of glioma patients at high risk of glioma (50 responses)
[0024] 1. RNA stabilization solution 50ml
[0025] 2. Isopropyl alcohol 100ml
[0026] 3. Trichloromethane 100ml
[0027] 4. Trizol 50ml
[0028] 5. 10ml of enzyme-free water
[0029] 6.1μM random reverse transcription primer 50ul
[0030] 7.5× reverse transcription buffer 200ml
[0031] 8.10mM triphosphate base deoxynucleotide 100ul
[0032] 9.40U / μl RNase inhibitor 500ul
[0033] 10.200U / μl MMLV reverse transcriptase 50ul
[0034] 11.PremixExTaq50ul
[0035] 12.10 μM ncRNAPRKAG2 real-time quantitative PCR specific primers 30ul
[0036] CASC2 forward primer 5′-TTGACCCTTCCAGCTTCC-3′,
[0037] CASC2 reverse primer 5'-CCATCCGCACATCACAAT-3',
[0038] 13.10μM 6snRNA specific primer 30ul
[0039] The forward primer is 5'-ATTGGAACGATACAGAGAAGATT-3',
[0040] The forward primer was 5'-GGAACGCTTCACGAATTTG...
Embodiment 2
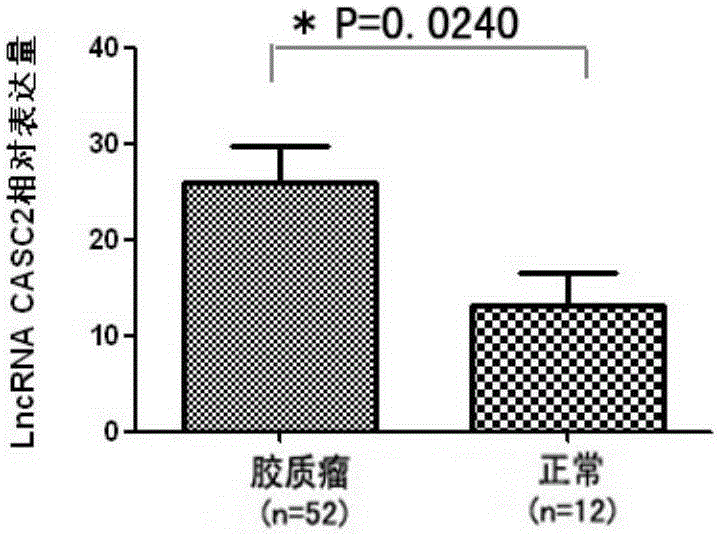
[0042] Validation of differential expression of LncRNACASC2 in glioma tissue and normal brain tissue
[0043] 1. Collect the glioma tissue to be tested, put it into a cryopreservation tube containing RNA stabilization solution, and store it in a -80°C refrigerator for later use.
[0044]2. Extraction of RNA from tissue: Take an appropriate amount of the sample and add liquid nitrogen to the mortar after baking at 180°C for 6-8 hours to grind the sample, grind it to a powder, add 1ml Trizol mortar sample to the mortar, and grind it into a liquid Transfer to a tube, add 200 μl / ml of chloroform Trizol to the tube, shake by hand for 15-30 s, place on ice for 5 min, and centrifuge at 12,000g at 4°C for 15 min; carefully take the upper aqueous phase into a new tube, add pre-cooled isopropyl alcohol Mix well with 0.5ml / ml Trizol of propanol, let stand at -20°C for 20min, centrifuge at 12000g for 10min at 4°C; discard the supernatant, add 1-2ml of ethanol diluted with 75% DEPC water, ...
Embodiment 3
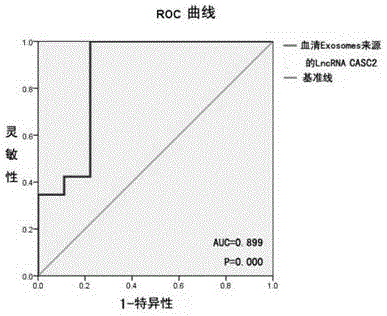
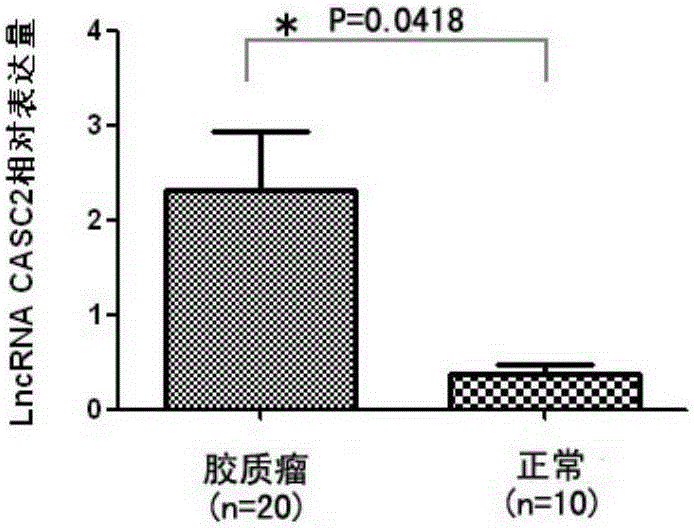
[0055] Specific and sensitive detection of serum Exosomes-derived LncRNACASC2 for glioma diagnosis
[0056] 1. Isolation of Exosomes in Serum
[0057] Collection of peripheral blood from individuals to be tested
[0058] 1.1 Separation of peripheral blood serum: use a blood coagulation tube to collect 5ml of blood from the individual to be tested. The blood was centrifuged at 1000 rpm for 6 min after blood collection, and the serum was drawn and stored in an EP tube at -80°C.
[0059] 1.2 Separation of Exosomes in serum: Add 100 μl of TotalExosomeIsolationReagent to 500 μl of each serum sample, mix by vortexing, and react at 4°C for 30 minutes. Centrifuge at 10,000g for 10 minutes at room temperature. There are Exosomes at the bottom of the EP tube, resuspend the Exosomes in 200 μl PBS. (Choose a commercial Exosomes isolation kit)
[0060] 2. Extraction and purification of RNA from Exosomes (select a commercialized Exosomes isolation and purification RNA kit)
[0061] 2....
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
| Sensitivity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com