A method for batch detection of ltr-retrotransposons in plant genomes
A technology of retrotransposon and batch detection, which is applied in special data processing applications, instruments, electrical digital data processing, etc., can solve the problem that the program cannot be identified, and achieves the effect of fast speed, good effect and easy process flow.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0042] Example 1. Establishment of a method for batch detection of plant genome LTR-retrotransposons
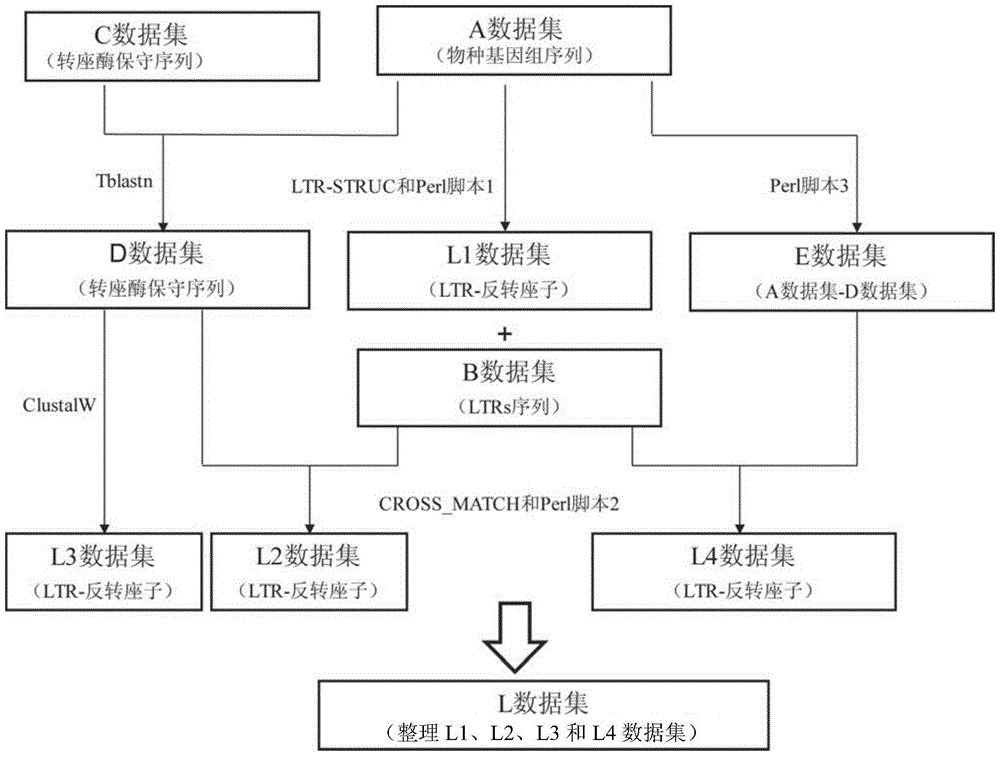
[0043] The flowchart of the method for batch detection of plant genome LTR-retrotransposons provided by the present invention is shown in figure 2 , including the following steps:
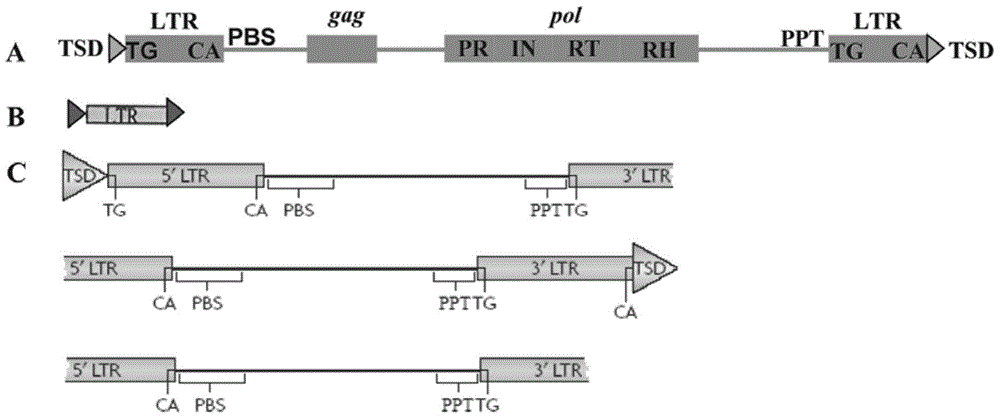
[0044] (1) Use the LTR_STRUC program based on the de novo search for the structural characteristics of transposons to identify relatively young LTR-retrotransposons in the genome to be tested (data set A), operate under the Windows system, and use the default parameter settings for analysis; then , using Perl script language programming (Script 1), follow the steps below to extract LTR-retrotransposon sequences (L1 data set) and LTRs sequences (B data set);
[0045] Extraction of LTR-retrotransposons and LTR sequence steps: put the script 1 under the LTRSTRUC folder, run the "perl abstract1.pl XXX1" command, where "XXX1" represents the file name of the A data set (for LTR_STRUC analysis has bee...
Embodiment 2
[0073] Example 2, using the method established in Example 1 to detect the Raymond cotton genome LTR-retrotransposon in batches
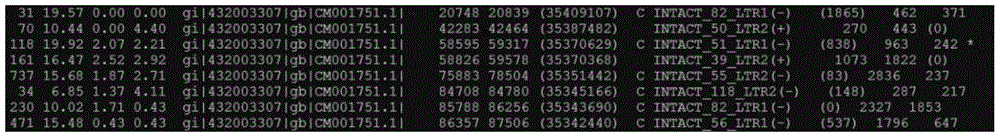
[0074] Enter the NCBI database (BioProject accession PRJNA171262) to download the genome sequence (13 chromosomes, 737.8Mb) of Gossypium raimondii L., upload it to the Windows system and the local Linux computing server, and perform LTR of the Gossypium raimondii L. genome - Detection of retrotransposons. During the detection process, the names, operating environments and addresses of commonly used programs involved are shown in Table 1. The specific operation steps of the detection method are as follows:
[0075] 1) Carry out with reference to step (1) of Example 1.
[0076] The LTR_STRUC program was used to identify LTR-retrotransposons in the Raymond cotton genome (Dataset A), operating under the Windows system with default parameter settings. Using Perl script language programming (script 1), extract 2247 LTR-retrotransposon sequence sets (L1 ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com