Gluconobacter oxydans promoter and its application
A technology for oxidizing glucose and acid bacteria, which is applied in the field of promoters, can solve the problems of lack of constraints on the genetic engineering improvement of Gluconobacter oxidans, and achieve the effect of high activation activity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0038] Example 1: Cloning of the promoter gHp0169
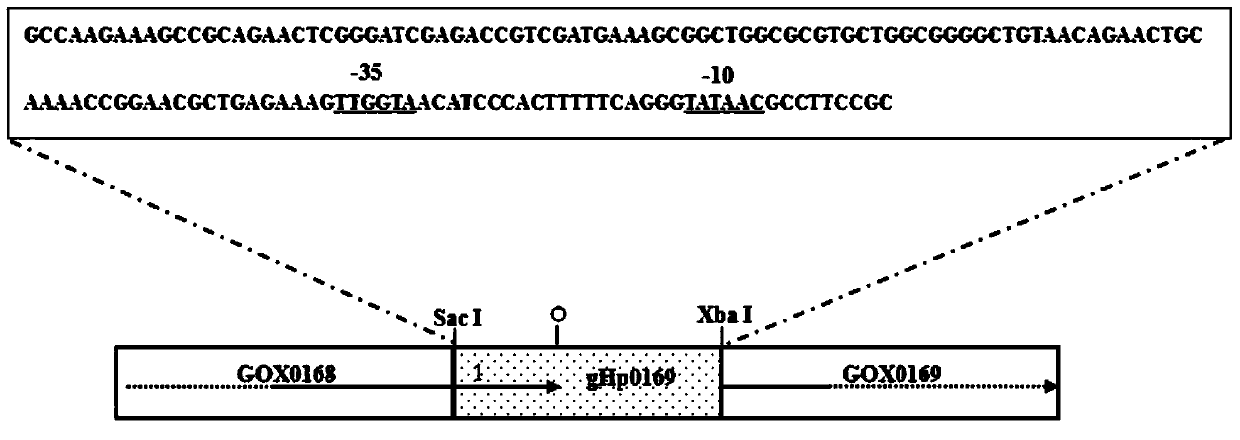
[0039] According to the G.oxydans621H genome sequence, the gHp0169 sequence is located between GOX0168 and GOX0169 (eg figure 2 shown), design and synthesize primers (Shanghai Jierui Bioengineering Co., Ltd.), its sequence is as follows:
[0040] gHp0169-F: 5'-ATA GAGCTC GCCAAGAAAGCCGCAGAACTC-3';
[0041] gHp0169-R: 5'-GCA TCTAGA GCGGAAGGCGTTATACCCTGA-3';
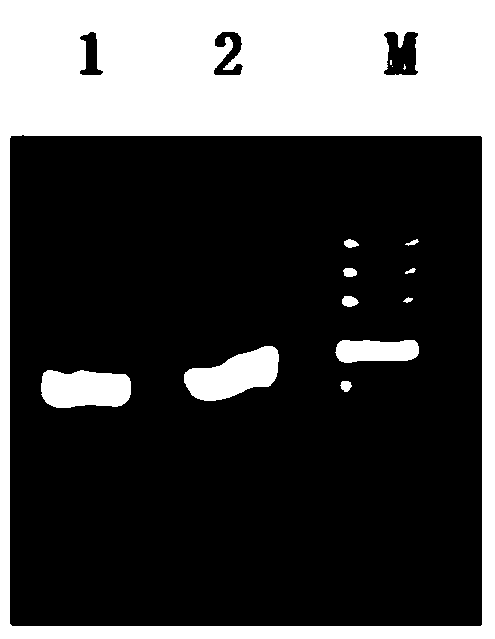
[0042] Using the genome of G.oxydans as a template, using gHp0169-F and gHp0169-R as primers, PCR amplifies the target gene, and introduces Sac I and Xba I restriction sites (underlined) at both ends, respectively. The specific conditions are as follows :
[0043] Reaction system: 2×PCR mix 25μl, primer (10μmol / L) 1μl each, template 1μl, ddH 2 O22μl;
[0044] Reaction process: 94°C for 5min, 94°C for 30s, 60°C for 30s, 72°C for 30s, 30 cycles, 72°C extension for 10min, 4°C storage.
[0045] The result is as figure 1 As shown, primers gHp0169-F and gHp0169-R wer...
Embodiment 2
[0047] Example 2: Verification of the functional activity of the promoter gHp0169
[0048] 2.1. Construction of plasmid pBBR1pgHp0169-GFP
[0049] According to the enzyme cutting sites designed at both ends of the primers gHp0169-F and gHp0169-R, the gHp0169 fragment was excised from the T-gHp0169 recombinant vector constructed in Example 1 with Sac I and XbaI, and the recovered gHp0169 fragment was Ligated with pBBR1MCS5 vector fragment digested with Sac I and Xba I. The ligated product was transformed into Escherichia coli DH5α strain, the plasmid was extracted, and identified by digestion with Sac I and Xba I. Sequencing was performed on the initially identified correct recombinant plasmid, and the recombinant vector containing 142 bp shown in SEQ ID NO: 1 was named pBBR1pgHp0169.
[0050] Using PET28a-GFP (purchased from Luyang Biopharmaceutical Company, USA) as a template, GFP primers (Shanghai Jierui Bioengineering Co., Ltd.) were designed and synthesized. The sequence...
Embodiment 3
[0083] Embodiment 3: Utilize promoter gHp0169 to overexpress ga2dh gene to improve 2-KGA production
[0084] 3.1. Construction of plasmids pBBR1pgHp0169-ga2dh and pBBR1ptufB-ga2dh
[0085] According to the nucleotide sequence of ga2dh of Gluconobacter oxydans G.oxydans, design primers (Shanghai Jierui Biological Engineering Co., Ltd.), the sequence is as follows:
[0086] ga2dh-F: 5'-CTA TCTAGA GGAGAAACCTGTGCCCCCCATG-3';
[0087] ga2h-R: 5'-GAG GGATCC TTCAGTTCAGTGAGACCGCATCATC-3';
[0088] Using the genome of G.oxydans as a template, using ga2dh-F and ga2dh-R as primers for PCR amplification, respectively introducing Xba I and BamH I restriction sites at both ends of the promoter sequence, the amplification conditions are as follows:
[0089] Reaction system: 2×PCR mix 25μl, primer (10μmol / L) 1μl each, template 1μl, ddH 2 O22μl;
[0090] Reaction process: 30 cycles of 94°C for 5min, 94°C for 30s, 59°C for 30s, 72°C for 4min, extension at 72°C for 10min, and storage at ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com