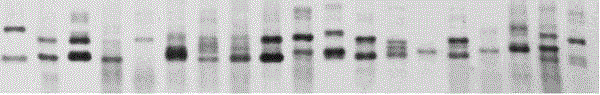
Camellia oleifera ssr molecular marker
A molecular marker, Camellia oleifera technology, applied in the field of development of Camellia oleifera microsatellite genetic markers, can solve the problems of little research on the development and utilization of Camellia oleifera microsatellite molecular markers, and achieve high reliability and good repeatability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0083] 1. Preparation method of camellia oleifera SSR molecular marker
[0084] 1.1 Acquisition and pretreatment of Camellia oleifera DNA sequence
[0085] Using the method of our research group (refer to JiangCetal. Efficient extraction of RNA from various Camellia species rich in secondary metabolites for deep transcriptome sequencing and gene expression analysis. African Journal of Biotechnology, 2011, 10 (74): 16769-16773.) to extract Camellia oleifera DNA and RNA, respectively construct genome and transcriptome 4 GS4 high-throughput libraries by FLX, respectively. Genome and EST sequences were obtained from the sequencing reaction. The EST sequence was spliced by CAP3 after removing low-quality sequences by SeqClean and Lucy software, and the genome sequence obtained by sequencing was processed and spliced by the Newbler software that comes with the 454 sequencer.
[0086] 1.2 Screening of SSR loci in Camellia oleifera DNA sequence
[0087] The SSR sites in each DNA...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com