Rapid Screening Method for Salmonella and Shigella in Enterobacteriaceae
A technology for Salmonella and Shigella, applied in the direction of microorganism-based methods, biochemical equipment and methods, and microorganism measurement/inspection, can solve the problems of high cost, complicated experimental methods, time-consuming biochemical tests, etc., and achieve low cost , The experimental method is simple, the effect of less substrate
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
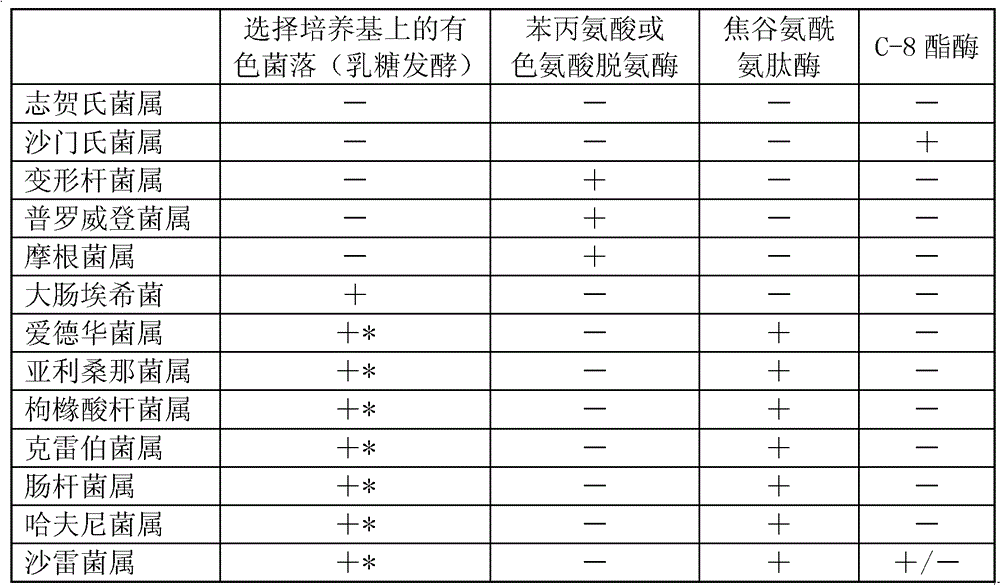
[0016] Example 1 of the present invention: Streak the specimen on a MacConkey agar plate, incubate at 37°C for 18 hours, then check the colorless colony growing on the plate; pick out a colorless colony, and use a part of it to detect oxidase activity; another part was inoculated on filter paper strips soaked with tryptophan, pyroglutamyl-4-methoxynaphthylamine and 4-methylumbelliferyl ioxynil caprylate and wetted with a drop of water or buffer . The appropriate amount of soaking liquid for filter paper is: 0.2ml of each of tryptophan, pyroglutamyl-4 methoxynaphthylamine and 4-methylumbelliferone ioxynil octanoate is soaked on 1 square inch of filter paper. 100 micrograms of 20 mM Tris buffer (pH 7.5), the filter paper used was Whatman No. 3 (Whatman Ltd. Maidstone England). The wetted inoculated strips were placed in a moisturizing vessel and incubated at 37°C for 2 hours, and then the filter paper strips were placed under a Wood lamp to detect fluorescence. Positive fluores...
Embodiment 2
[0017] Example 2 of the present invention: Streak the specimen on a MacConkey agar plate, incubate at 36°C for 20 hours, then check the colorless colony growing on the plate; pick out a colorless colony, and use a part of it to detect oxidase activity; another part was inoculated on filter paper strips soaked with tryptophan, pyroglutamyl-4-methoxynaphthylamine and 4-methylumbelliferyl ioxynil caprylate and wetted with a drop of water or buffer . The appropriate amount of soaking liquid for filter paper is: 0.3ml of each of tryptophan, pyroglutamyl-4 methoxynaphthylamine and 4-methylumbelliferone ioxynil octanoate is soaked on 1 square inch of filter paper. 160 μg of 20 mM Tris buffer (pH 7.5), the filter paper used is Whatman No. 3. The wet inoculated strips were placed in a moisturizing container and incubated at 36°C for 3 hours, and then the filter paper strips were placed under a Wood lamp to detect fluorescence. Positive fluorescence indicates the presence of C-8 estera...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com