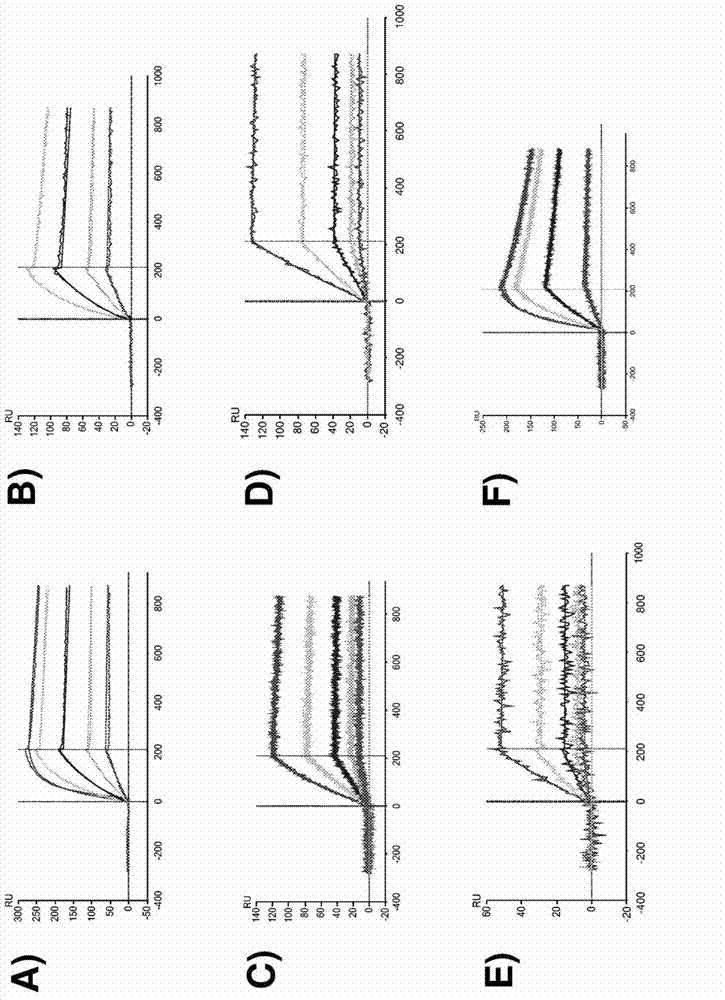
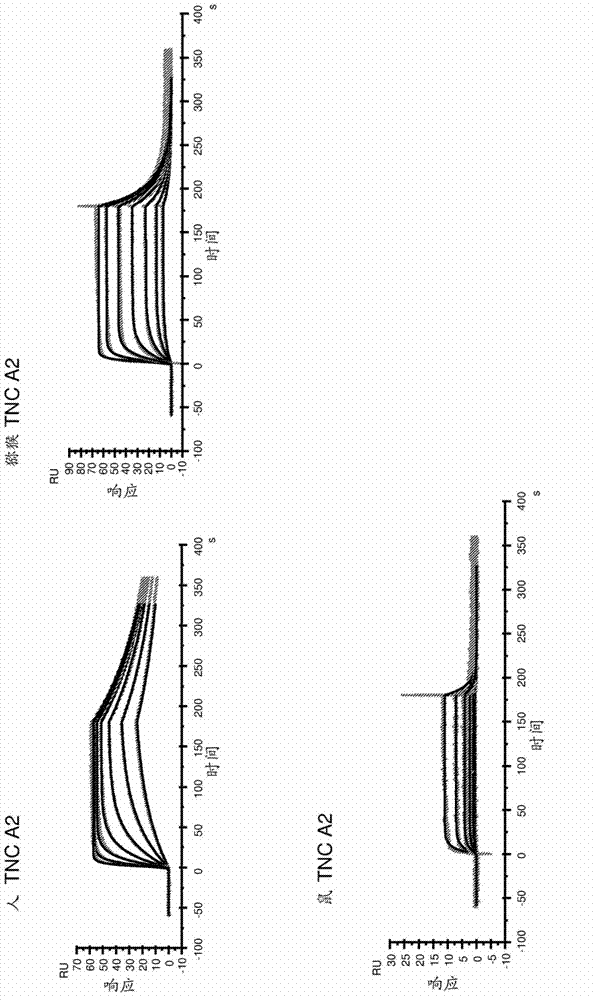
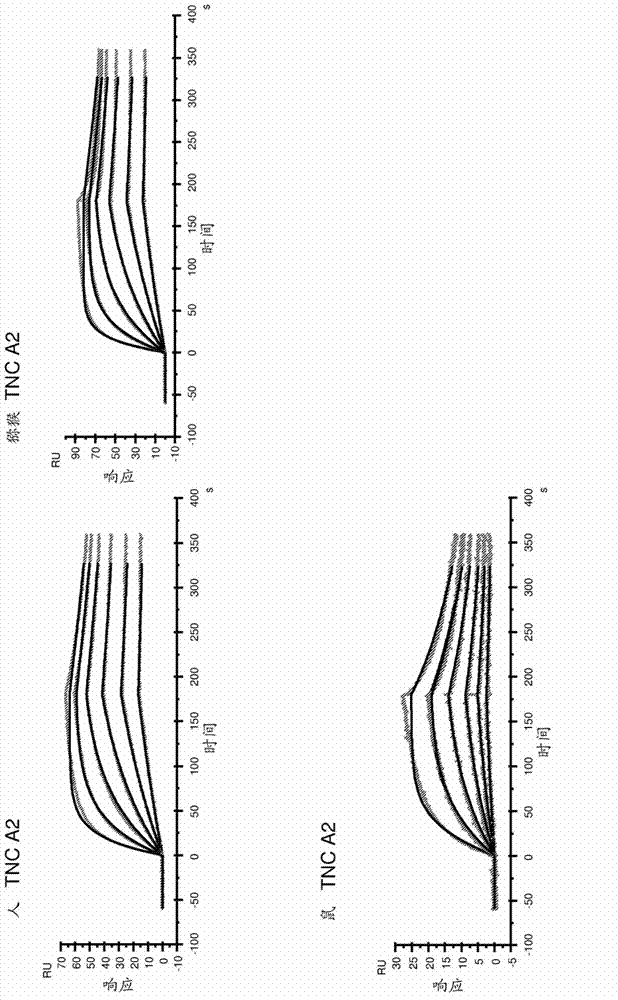
Anti-tenascin-c a2 antibodies and methods of use
A tenascin and antibody technology, applied in the direction of antibodies, anti-animal/human immunoglobulins, immunoglobulins, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0290] recombinant DNA technology
[0291] DNA was manipulated using standard methods, as described in Sambrook, J. et al., Molecular cloning: A laboratory manual; Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York, 1989. Molecular biology reagents were used according to the manufacturer's instructions. DNA sequences were determined by double-strand sequencing. In some cases, desired gene segments were prepared from synthetic oligonucleotides and PCR products by automated gene synthesis by Geneart AG (Regensburg, Germany). The gene segment flanked by unique restriction endonuclease cleavage sites was cloned into the pGA18(ampR) plasmid. Plasmid DNA was purified from transformed bacteria and concentrations were determined by UV spectroscopy. The DNA sequence of the subcloned gene fragment was confirmed by DNA sequencing. Gene segments were designed with appropriate restriction sites to allow subcloning into corresponding expression vectors.
[0292]For gener...
Embodiment 2
[0306] Construction of Universal Fab Libraries
[0307] Construction of a universal antibody library in Fab format based on human germline genes using the following V domain pairings: Vk3_20 kappa light chain and VH3_23 heavy chain for the DP47-3 library, and Vk1_17 kappa light chain and VH1_69 heavy chain for DP88 -3 library. See SEQ ID NOs 1 and 2.
[0308] Both libraries were randomized in the light chain CDR3 (L3) and heavy chain CDR3 (H3), and each library was assembled from 3 fragments by splicing by overlapping extension (SOE) PCR. Fragment 1 contains the 5' end of the antibody gene, including the randomized L3, Fragment 2 is the central constant fragment spanning L3 to H3, and Fragment 3 contains the randomized H3 and the 3' portion of the antibody gene.
[0309] The following primer combinations were used to generate library fragments of the DP47-3 library: Fragment 1 (LMB3-LibL1b_new), Fragment 2 (MS63-MS64), Fragment 3 (Lib2H-fdseqlong). See Table 3. The followi...
Embodiment 3
[0318] Selection of anti-TNC A2 clone 2B10 (primary selection)
[0319] Subclones were selected against human TNC-A2 expressed in E. coli at 5' of the avi tag and 6xhis tag. See SEQ ID NO:97.
[0320] Antigens are biotinylated in vivo upon expression. Select in solution according to the following scheme: (i) about 10 12 A library of DP88-3 phagemid particles and 100 nM biotinylated human TNC A2 were combined for 0.5 hours in a total volume of 1 ml; (ii) by adding 5.4x10 7 Streptavidin-coated magnetic beads for 10 minutes to capture biotinylated human TNC-A2 and attached phage; (iii) wash the beads with 5x1ml PBS / Tween20 and 5x1ml PBS; amine) for 10 min to elute the phage particles and neutralize by adding 500 μL 1M Tris / HCl pH 7.4; and (v) reinfect log-phase E. coli TG1 cells with helper phage VCSM13, followed by PEG / NaCl precipitation of the phage Bacteroid pellets for subsequent rounds of selection.
[0321] Selection was performed over 3 rounds using a constant antigen...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com