Hierarchical modeling method of protein side chain prediction
A modeling method and hierarchical technology, applied in special data processing applications, instruments, electrical digital data processing, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
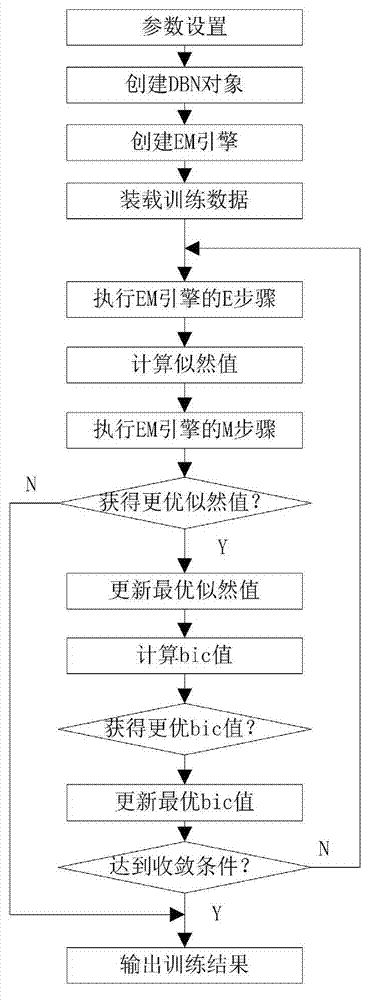
[0062] The training data used is the same 850 proteins as the training set used by the Dunbrack library. These data were obtained by X-ray crystallography in the PDB library. If the PDB structure contains multiple chains, only the A chain is selected, and the similarity between any two chains is guaranteed not to exceed 50%.
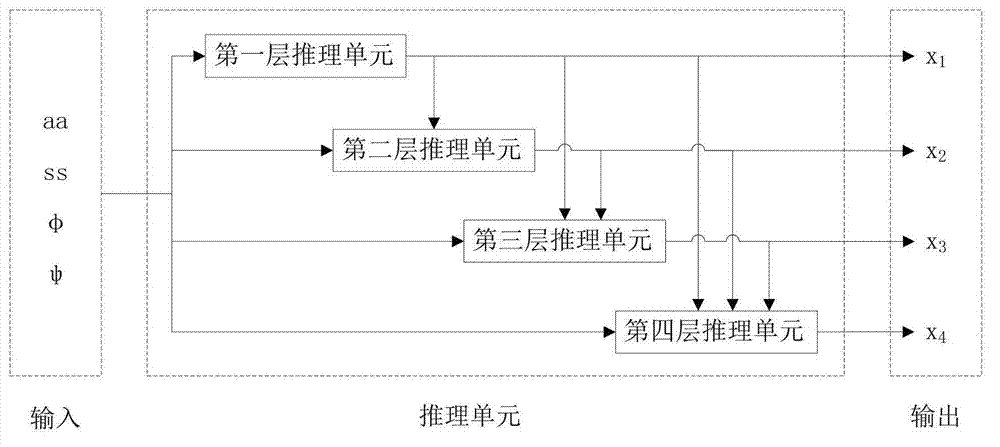
[0063] The training process requires the following 8 types of data: residue type aa, secondary structure type ss, main chain torsion angle φ, main chain torsion angle ψ, side chain torsion angles x1, x2, x3, and x4. Before training, it is necessary to digitize the information and discretize the continuous data. The first is the protein type. At present, there are only 20 common amino acids involved in protein composition. Therefore, the protein type itself is discretized information, which is represented by integers 0-19. Similarly, the secondary structure of the main chain is also discrete information. There are only three kinds of protein secondary ...
Embodiment 2
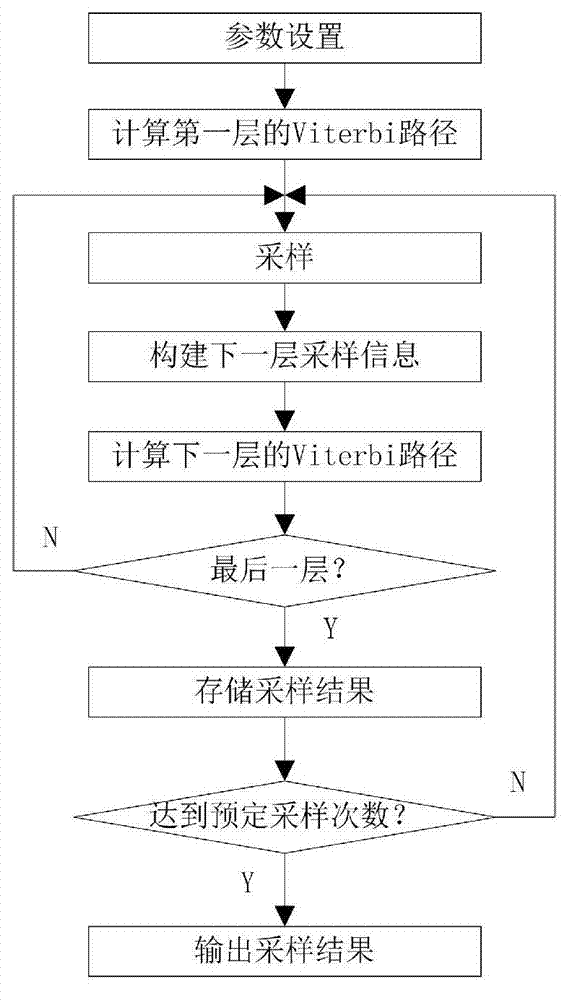
[0072] The present invention employs three GPCR targets: 2RH1, 3EML and 3OE6. For these three experimental data, the hierarchical modeling method proposed by the present invention and the traditional non-hierarchical modeling method are respectively used to generate corresponding rotamer libraries, and then the quality of the rotamer libraries is compared. The comparison method is to count the number of angles whose x-angle difference between the residue candidate structure at a certain position in different rotamer libraries and the corresponding position in the natural structure is within 20°. As shown in table 2.
[0073] Table 2 Sampling results of hierarchical modeling method
[0074]
[0075]Among them, the two columns of x1 and x2 represent the statistical results for the corners of x1 and x2 respectively; x1+x2 represents the statistical results when x1 and x2 are satisfied at the same time. "Four layers" means the experimental results of the rotamer library gener...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com