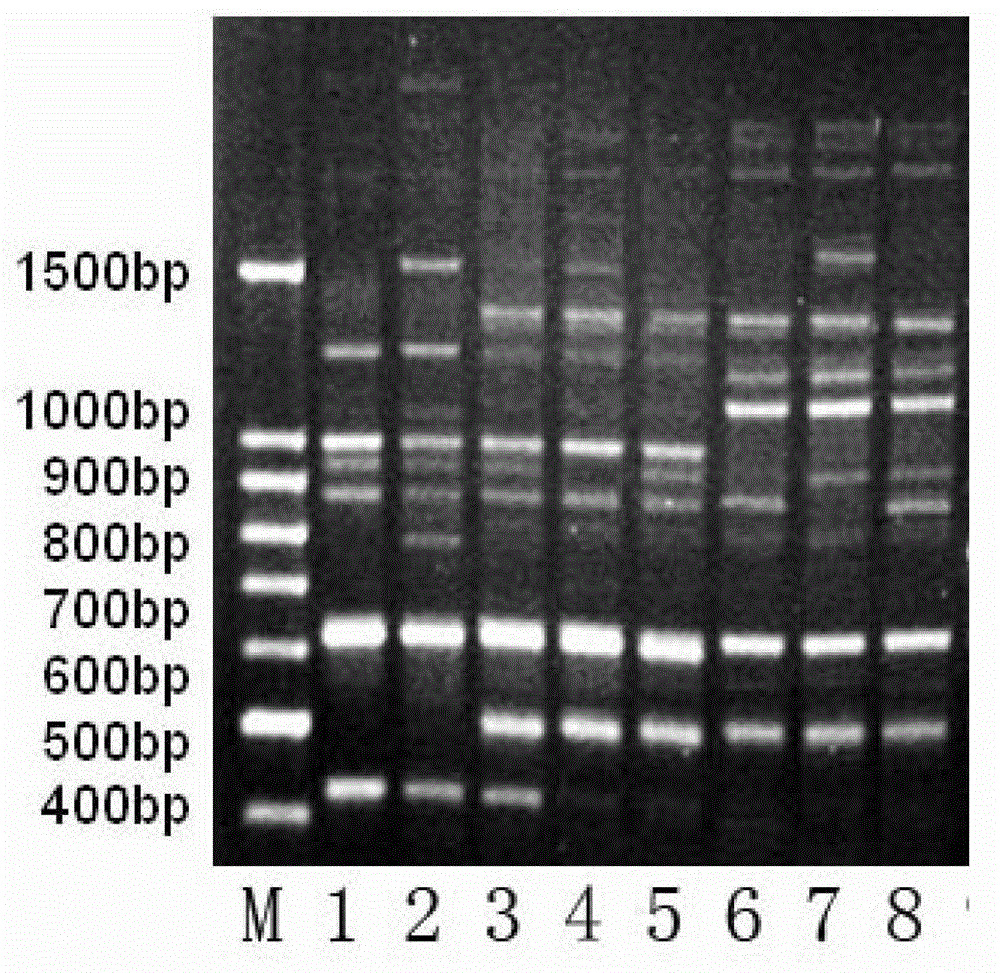
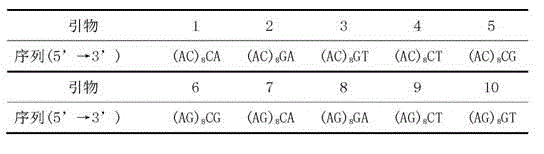
Molecular marker fingerprinting for identifying anoectochilus formosanus produced at Huoshan in Anhui and similar species thereof
A technology of fingerprints and molecular markers, which is applied in the field of fingerprints of Aurantia clematis, can solve problems such as time-consuming, high cost, and complicated procedures, and achieve the effect of simple steps, less sampling, and strong genotyping ability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
Embodiment
[0049] (1) Pretreatment of fresh clematis samples
[0050] Keep fresh clematis samples away from light for more than 6 hours to consume starch, take 10 grams of fresh clematis leaves, soak them in more than 50 grams of absolute ethanol twice, each time for 10 minutes, and wipe dry;
[0051] (2) Extraction of sample genomic DNA
[0052] The improved CTAB method was used to extract genomic DNA from leaves of A. clematis, and the operation was as follows:
[0053] 1) Put the pretreated leaves into a frozen ceramic mortar, add an appropriate amount of quartz sand, then add liquid nitrogen several times to fully grind to powder, and quickly transfer the leaf powder to a 10ml pre-cooled Eppendorf centrifuge tube with a medicine spoon When the height of the powder reaches 1 / 3 of the centrifuge tube, stop adding the powder;
[0054] 2) Add 4ml of 65°C preheated CTAB extraction buffer [2% (v / w) CTAB, 1.4M NaCl, 20mM EDTA, 100mM Tris-HCl (PH=8.0), 2% mercaptoethanol] into the centrifu...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com