Oligonucleotide library classification and assessment method based on capillary zone electrophoresis
An oligonucleotide library and zone electrophoresis technology, applied in the field of biological separation and analysis, can solve the problems of long time, heavy workload, reduce the time and operation steps of the screening process, and achieve the effect of simple data processing method.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
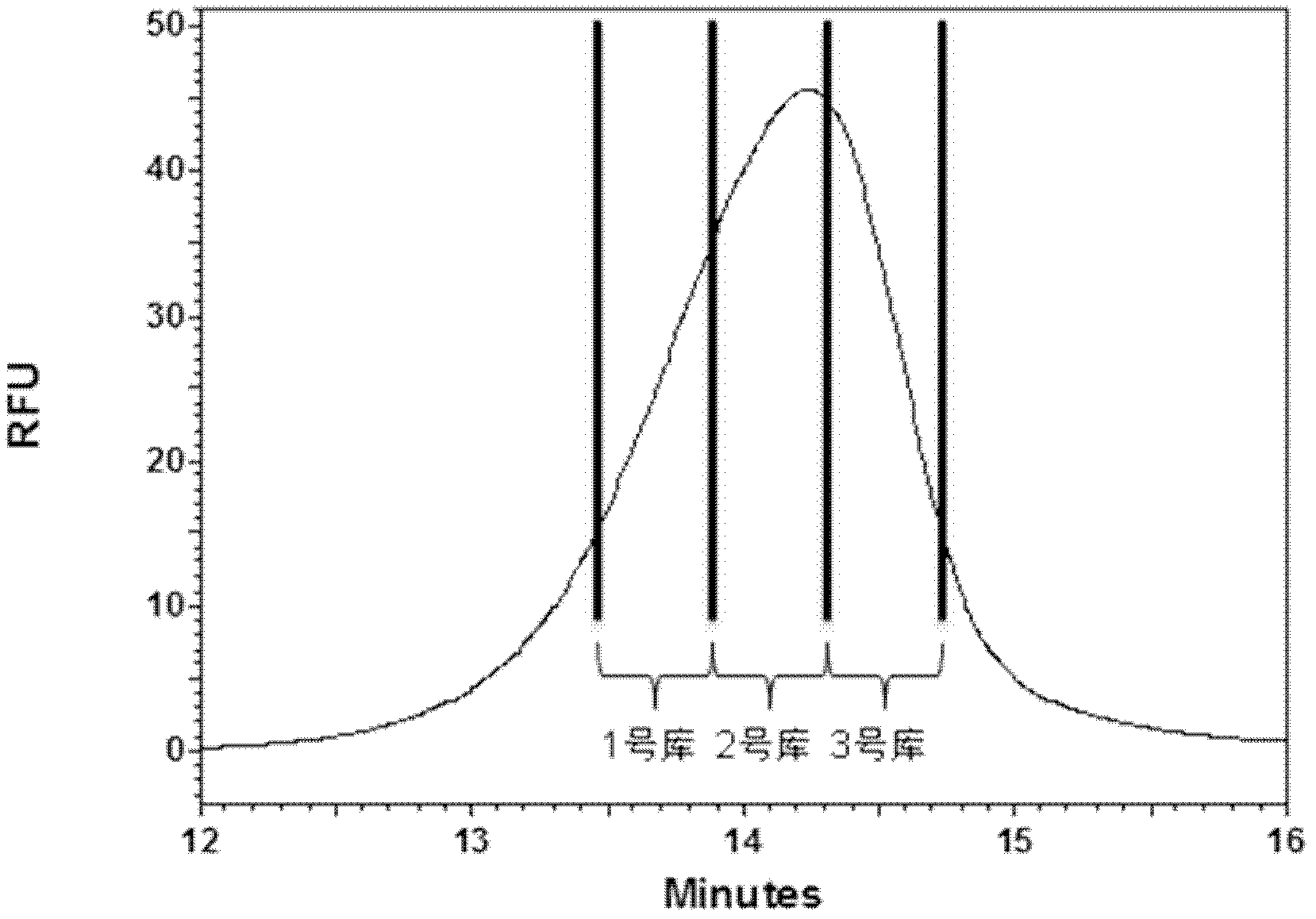
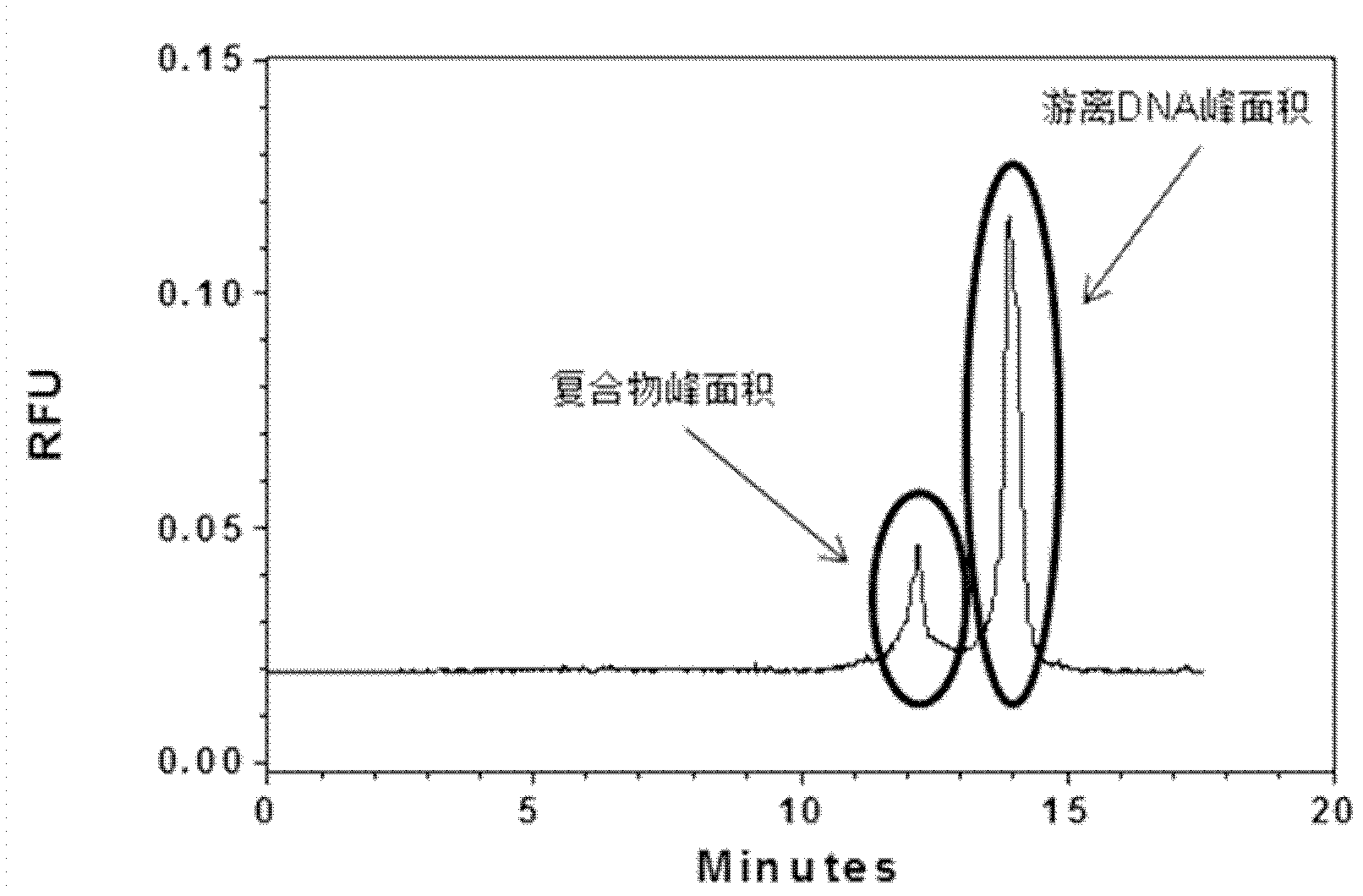
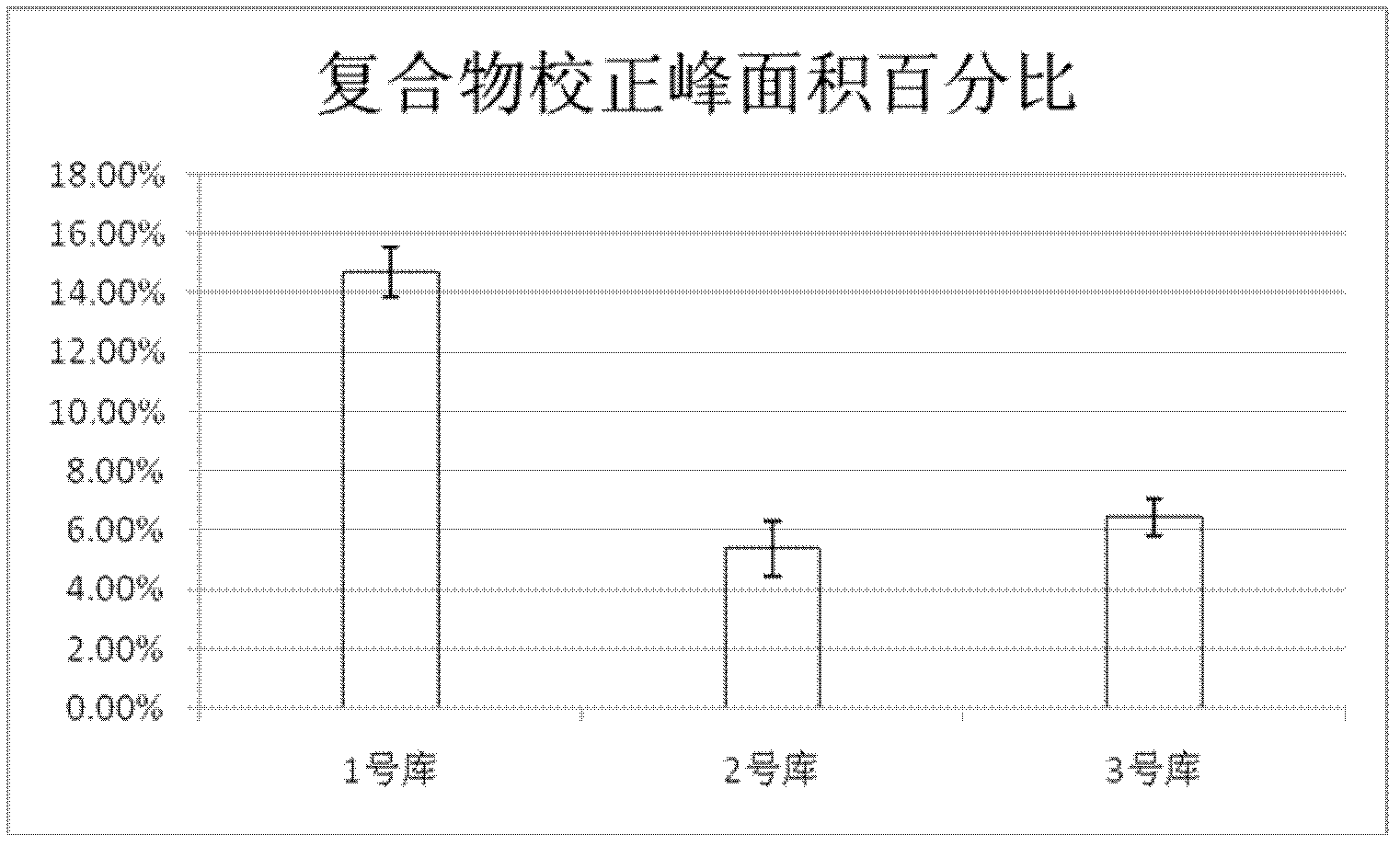
[0039] A method for grading and evaluating an oligonucleotide library based on capillary zone electrophoresis, the steps are as follows:
[0040] step one,
[0041] (1) Dissolve the oligonucleotide library in the running buffer solution to make a stock solution 1 with a concentration of 100 μM of the oligonucleotide library, and store it at -20°C for later use; then dilute the stock solution 1 into a stock solution with a concentration of 20 μM stock solution 2;
[0042] Among them, the capillary used for capillary zone electrophoresis is a quartz fused capillary, which is covered with a silica gel layer. The inner diameter of the capillary is 75 μm, the total length is 50.2 cm, the side length of the detection window is 0.2 cm, and the distance from the window to the sample outlet is 10 cm; Rinse with water and running buffer solution for 3 minutes each; after washing, transfer the stock solution 2 to the capillary zone electrophoresis sample bottle, and use pressure injecti...
PUM
| Property | Measurement | Unit |
|---|---|---|
| The inside diameter of | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com