Heat-resisting beta-1, 3-1, 4-dextranase and encoding gene thereof
A coding gene and gene technology, applied in the field of β-1, can solve the problems of large differences in glucanase activity and achieve the effects of high expression level, heat resistance characteristics of expression level, and great potential for popularization and application
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0042] Example 1, Discovery of β-1,3-1,4-glucanase gene
[0043] 1. PCR amplification of the conserved region of the glucanase gene genome
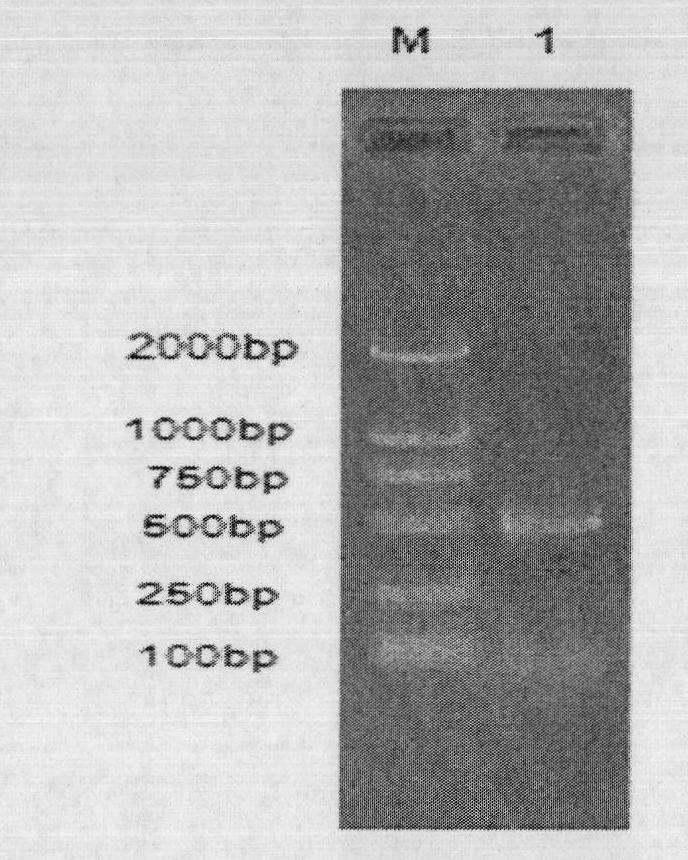
[0044] With the genomic DNA of Paecilomyces thermophila J18 (CAS Microbiology Institute Culture Collection Center, preservation number CGMCC 6885) as a template, according to the amino acid sequence of the filamentous fungal β-glucanase reported by GeneBank, Using the online software Block Maker( http: / / blocks.fhcrc.org / blocks / blockmkr / make blocks.html ) to search for the conserved region, and then use the online primer design software CODEHOP (http: / / blocks.fhcrc.org / codehop.html) to design the degenerate primer PtLic16CP1 (forward): 5′-CGGCGAGATCGACATCATHGARGGNGT-3′, PtLic16CP2 (reverse) : 5'-GCCCAGTCGCGCARAANGTNRRTC-3'. PCR reaction mixture (50 μL): water 37.25 μL; 10×Ex Taq buffer, 2.5 μL; 2.5 mmol / L dNTPs, 4 μL; primers (10 μmol / L), each 2.5 μL; DNA template (50-100ng), 1 μL; Taq (5U / μL), 0.25 μL. PCR reaction conditions: 94°C f...
Embodiment 2
[0047] Embodiment 2, expression of recombinant β-1,3-1,4-glucanase gene
[0048] 1. Construction of recombinant bacteria
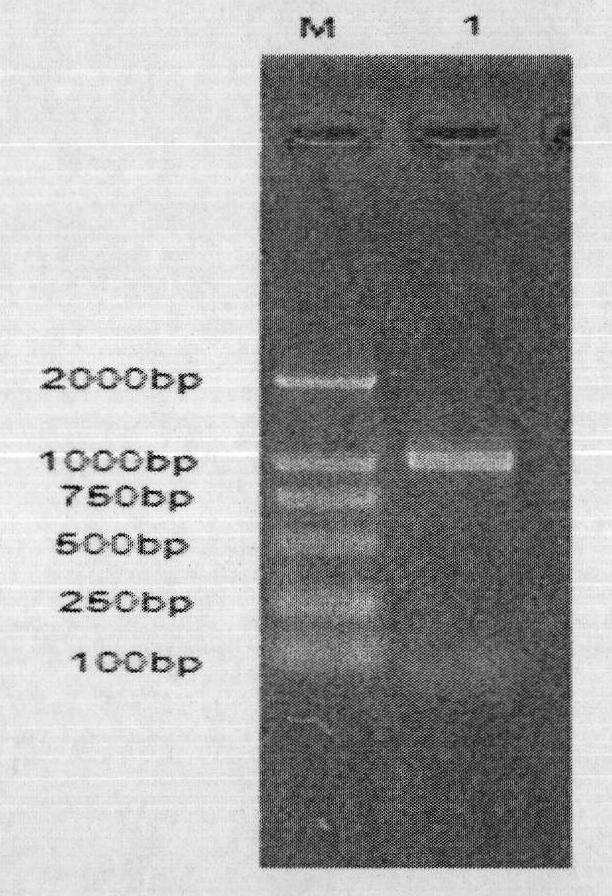
[0049] The complete open reading frame (ORF) encoded protein sequence was analyzed by SignalP 3.0, and there was a signal peptide with a possible cleavage site between amino acids 18 and 19 (AAA-YH). Based on this, primers were designed to remove the stop codon of the original gene. The two pairs of primers were: PtLic16ASnaBIF: 5′-TACGTATATCATCTTGTTGACGACTACG-3′ (including SnaBI restriction site); PtLic16AAvr II R: 5′- CCTAGG TTAGGGTGCGTACACGCGGAG-3' (containing the Avr II restriction site) was amplified by PCR using the above primers and the cDNA of Paecilomyces thermophila (Paecilomyces thermophila) J18 as a template. Agarose gel electrophoresis of PCR amplification products figure 2 After being recovered by the gel kit, it was cloned into the pMD-18T vector by TA cloning, transformed into Escherichia coli JM109 by heat shock method, and the recomb...
Embodiment 3
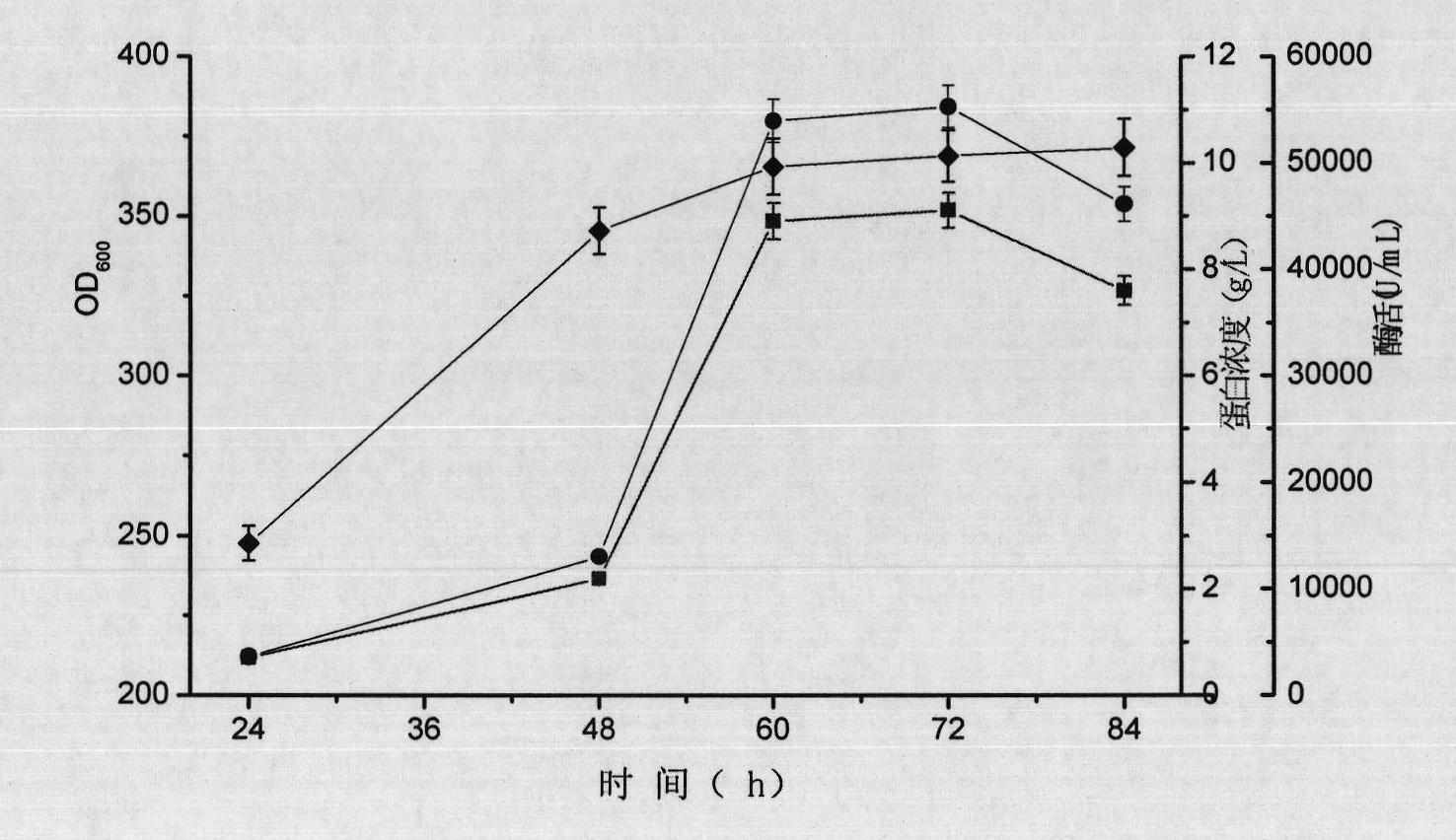
[0074] Embodiment 3, beta-1,3-1, the property of 4-glucanase
[0075] 1. Optimal pH of β-1,3-1,4-glucanase
[0076] The purified protein in Example 2 was dissolved in 4 different 50mM buffer systems (Citric-Na 2 HPO 4 , pH 2.5-5.5; MES, pH 5.0-6.5; phosphate buffer, pH 6.5-8.5; Glycine-NaOH, pH 8.5-11.0), and then measure the enzyme activity at 70°C (the enzyme activity determination method is the same as step 2) , taking the highest point of enzyme activity as 100% for plotting. The results show that the optimum pH of recombinant β-1,3-1,4-glucanase is 7.0 ( Figure 6 ).
[0077] 2. Optimum reaction temperature of β-1,3-1,4-glucanase
[0078] The purified protein in Example 2 was appropriately diluted in 50mM MES buffer solution with pH 7.0, and then the activity of β-1,3-1,4-glucanase was determined according to the above method at different temperatures of 40-100°C. Enzyme activity (the enzyme activity assay method is the same as step 2). The highest point of enzyme ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com