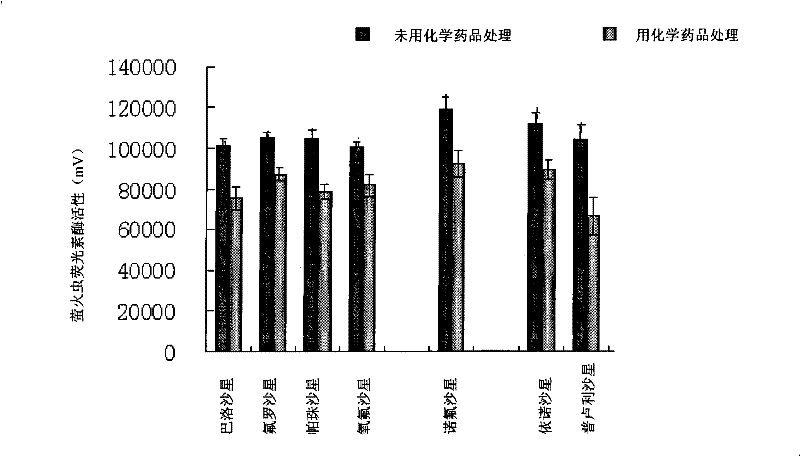
Composition for suppressing target gene expression
A technology for targeting genes and compositions, applied in gene therapy, drug combinations, active ingredients of heterocyclic compounds, etc., can solve the problems of reduced efficacy, off-target, low siRNA efficacy, etc., to achieve the effect of improving efficacy and reducing off-target
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0065] This example was used to determine the activity of siRNA alone and in combination with fluoroquinolones.
[0066] (1) Cell culture and transfection
[0067] Human embryonic kidney cells (HEK-293) were incubated at 37°C and 5% CO 2 They were cultured in DMEM / HIGH GLUCOSE (GIBICO) supplemented with 10% fetal bovine serum (FBS), 100 U / mL penicillin, 100 μg / mL streptomycin and 4 mM L-glutamic acid. Cells are maintained in exponential growth. Improved scheme (S.M.Elbashir, J.Harborth, W.Lendeckel, A.Yalcin, K.Weber, T.Tuschl, Duplexes of 21-nucleotide RNAs mediate RNA interference in culturedmammalian cells, Nature.411(2001) 494 by reported -498.), transfection of two luciferase plasmids. After transfection, cells were seeded into 24-well plates (0.5 ml medium / well) to reach confluence (approximately 50%) at the time of transfection. After culturing for 24 hours, the medium was replaced with OPTIMEM 1 (GIBICO) (0.5 ml / well) before transfection. Cells were co-transfected...
Embodiment 2
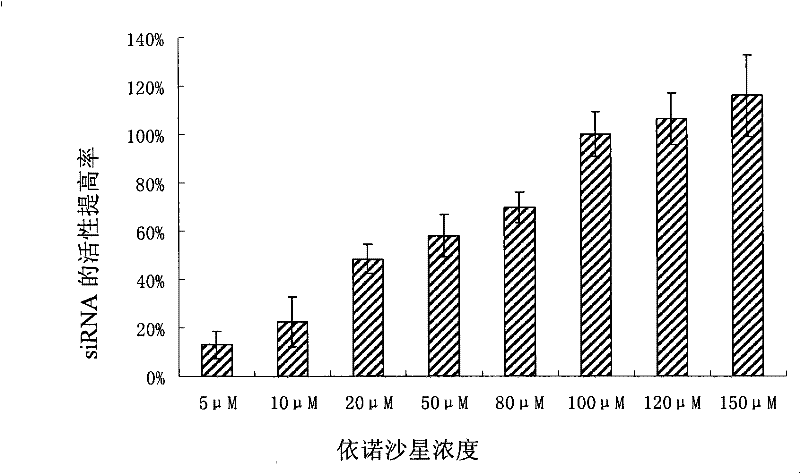
[0074] This example is to illustrate the relationship between the concentration of enoxacin and the improvement of siRNA activity.
[0075] Using different concentrations of enoxacin, HEK-293 was cultured and transfected according to the same method as in Example 1, and then the activity of luciferase was measured. The activity improvement rate of siRNA produced by 100 μM enoxacin was set at 100%.
[0076] From figure 2 It can be seen that the 50% effective concentration (EC50) of enoxacin is about 30 [mu]M.
Embodiment 3
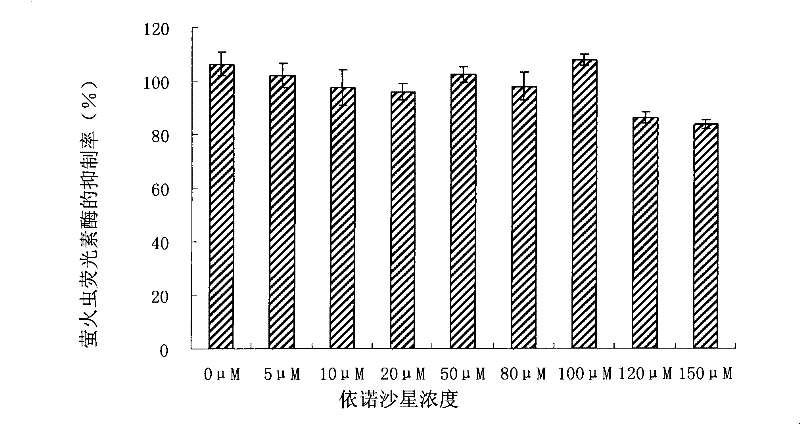
[0078] This example was used to determine the effect of enoxacin alone on firefly luciferase.
[0079] HEK-293 was cultivated and transfected in the same manner as in Example 1, except that only plasmids were used instead of siRNA.
[0080] Then, according to the same method as in Example 1, the cells were treated with different concentrations of enoxacin, and the activity of luciferase was determined. Inhibition by enoxacin was expressed as a normalized ratio between the luciferase reporter gene and the luciferase control gene. Luciferase control gene refers to samples not treated with the chemicals. The result is as image 3 shown.
[0081] From image 3 It can be seen that only at 120 μM and 150 μM, enoxacin inhibits the activity of luciferase, and has no effect on the activity of luciferase at concentrations lower than 120 μM. However, as measured in Example 2, the 50% effective concentration (EC50) of enoxacin is only about 30 μM. Therefore, within the effective concen...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com