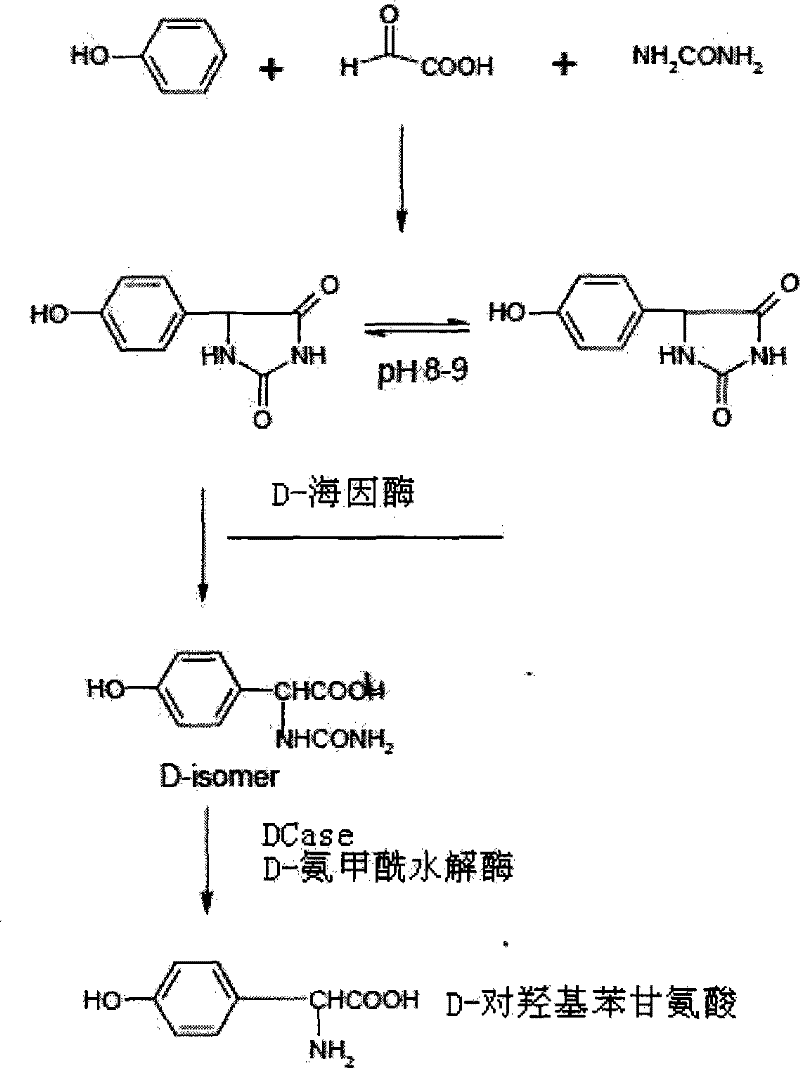
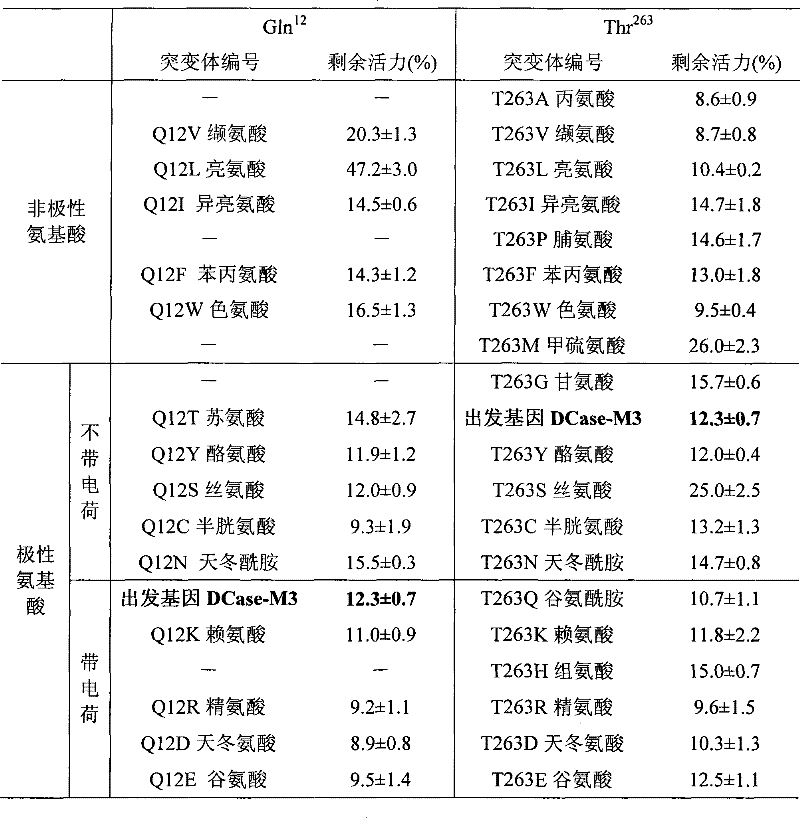
Mutant of D-carbamyl hydrolysis enzyme and application thereof
A kind of carbamoyl hydrolase and mutant technology, applied in the application of producing D-p-hydroxyphenylglycine, the field of mutants of D-carbamyl hydrolase, can solve the problems such as thermal stability is not improved
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0032] Example 1 , Cloning of DCase-M3 gene
[0033] 1.1, PCR amplification
[0034] Design a pair of primers:
[0035] 5'-GCTTTCAGAGTTCCGCGATCA-3'; and
[0036] 5'-TATGACACGTCAGATGATACTTGC-3'.
[0037] PCR amplification was performed using pET28b-Dcase-M3 (Jiang, S., et al., Biochem J, 2007.402(3): p.429-37) as a template.
[0038] PCR system: 17.75 μl redistilled water, 2.5 μl PCR buffer, 1 μl MgCl 2 (25 mM), 1.5 μl dNTP (2.5 mM), 0.5 μl of the above two primers, 15 ng of the above template, and 2 units of Taq enzyme (Shanghai Sangon Company).
[0039] PCR conditions: denaturation at 94°C for 10 min, denaturation at 94°C for 30 sec, annealing at 58°C for 30 sec, extension at 72°C for 50 sec, a total of 30 cycles, and finally full extension at 72°C for 10 min.
[0040] 1.2. Construction of plasmid containing DCase-M3 gene
[0041] After PCR amplification, first use 1% agarose gel to carry out nucleotide electrophoresis with a voltage of 100 volts. After cutting the gel...
Embodiment 2
[0046] Example 2 , Random mutation of DCase-M3 gene
[0047] Primer pair:
[0048] 5'-GCTTTCAGAGTTCCGCGATCA-3'; and
[0049] 5'-TATGACACGTCAGATGATACTTGC-3'.
[0050] Error-prone PCR reaction system (50μl): 20ng of template pET28b-Dcase-M3 (Jiang, S., et al., Biochem J, 2007.402(3): p.429-37), a pair of primers of 30pmol each, 7mM MgCl 2 , 50mM KCl, 10mM Tris-HCl (pH8.3), 0.2mM dGTP, 0.2mM dATP, 1mM dCTP, 1mM dTTP, 0.05mM MnCl 2 , and 5 units of Taq enzyme (Shanghai Sangon Company).
[0051] PCR reaction conditions: After denaturation at 94°C for 10 min, denaturation at 94°C for 30 sec, annealing at 58°C for 30 sec, extension at 72°C for 50 sec, a total of 30 cycles, and finally a full extension at 72°C for 10 min.
[0052] After PCR amplification, first use 1% agarose gel for nucleotide electrophoresis with a voltage of 100 volts. After cutting the gel, use the gel recovery kit (Shanghai Huashun Bioengineering Co., Ltd.) to recover the target DNA fragment of about 900 b...
Embodiment 3
[0057] Example 3 , screening of mutant libraries
[0058] 3.1. Transformation of mutants
[0059] The mutant constructed in Example 2 was transformed into E. coli DH10B (Invitrogen) by electric shock method. Transformed cells were smeared on LB plates (1% peptone, 0.5% yeast extract, 1% sodium chloride, 2% agar) containing ampicillin antibiotics, and cultured at 37°C.
[0060] 3.2 Mutant screening and identification
[0061] After the bacteria grew for 12 hours, a single colony was picked and added to a 96-well cell culture plate with 300 μl / well, 1 single colony / well, cultured on a shaker at 37° C. and 220 rpm. After 2 hours, the cells were induced with 1 mM IPTG, and cultured for 18 hours.
[0062] Take 100 μl / well of the cultured bacterial liquid into a 96-well microtiter plate, place the whole in a -70°C environment, and transfer it to a 37°C environment for recovery after 1.5 hours. The recovery time is about 30 minutes.
[0063] Mix the resuscitated bacterial solut...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com