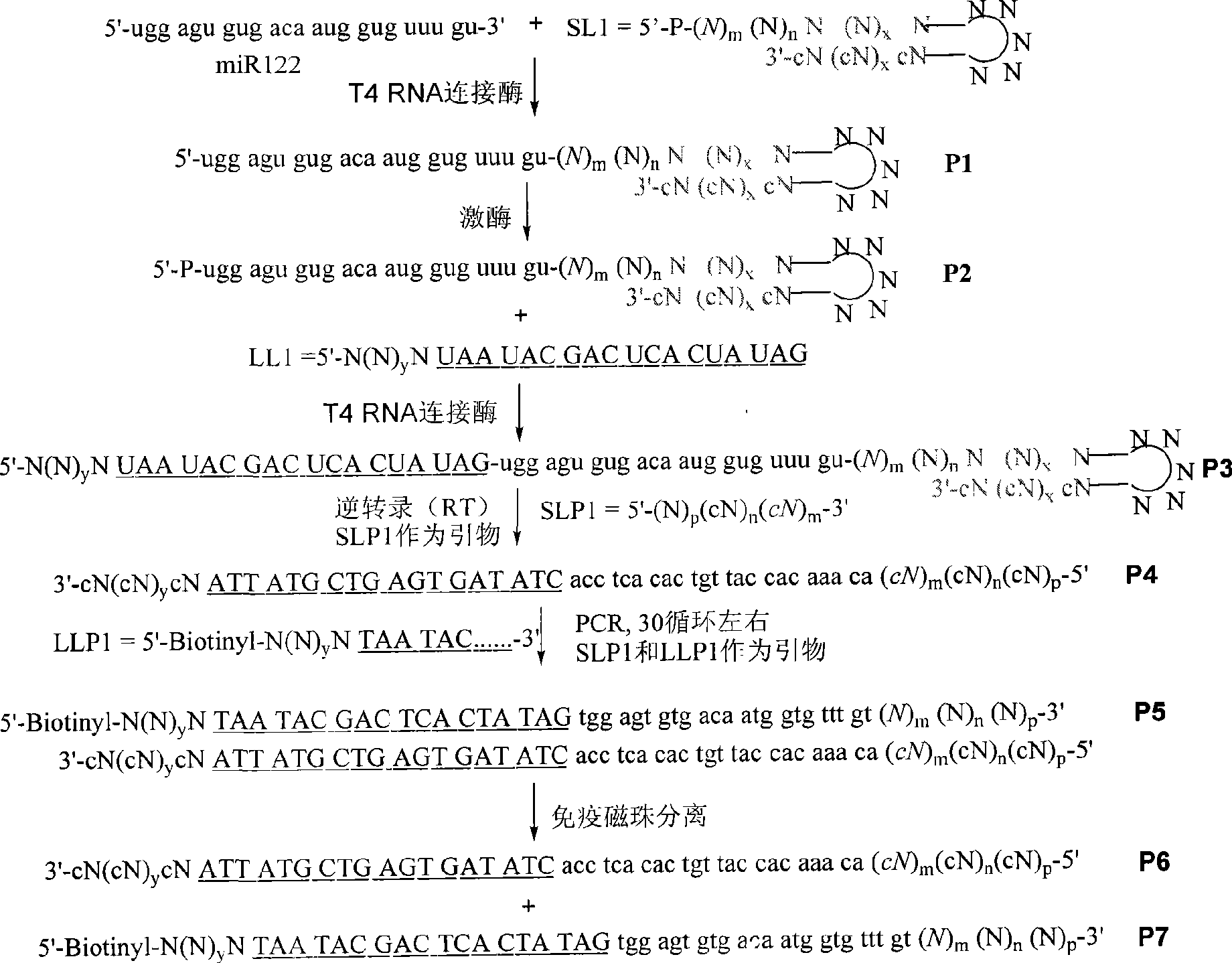Reagent used for ex vivo expansion and method thereof
A technology for in vitro amplification and reagents, applied in the reagents and fields for in vitro amplification of RNA, which can solve the problems of unknown RNA and RNA mixtures without amplification or effective detection
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0085] The experimental process of the restriction endonuclease cutting site at the 3' end of the recognition sequence is as follows (see figure 1 and figure 2 ). This example uses the restriction endonuclease NlaIII Electrophoresis results see Figure 4 .
[0086] A. Ligation of appropriate SL1 to target RNA, the resulting product is called P1.
[0087] In order not to be self-ligated, it is first necessary to dephosphorylate the 5' end of the target RNA, and then make the 5' phosphorylated SL1 under the action of a ligase such as T4 RNA ligase (NlaIII is used in the embodiments of the present invention, so SL1 The 5' end contains the site of endonuclease NIaIII) and dephosphorylated target RNA. SL1 can be DNA or RNA, or a DNA-RNA hybrid strand with RNA at the 5' end. Generally speaking, the ligation efficiency of RNA single strand to DNA single strand is lower than that of two RNAs. Since the 3' end of SL1 is blocked or in base pairing, T4 RNA ligase, which can only ...
Embodiment 2
[0108] The experimental process of the restriction endonuclease cutting site at the 5' end of the recognition sequence is as follows (see figure 1 and image 3 ). This embodiment uses restriction endonuclease DPNII, and its enzyme cutting site is: Electrophoresis results see Figure 5 .
[0109] Steps A-D are the same as in Example 1 except that the 3' end of SL1 contains a site for endonuclease DPNII.
[0110] E. Perform PCR on P4 to obtain P5.
[0111] Using SLP1 with free hydroxyl at the 5' end and LLP1 with biotin at the 5' end as primers or SLP1 with biotin at the 5' end and LLP1 with free hydroxyl at the 5' end, PCR amplification was performed on P4 to obtain the product P5. There are promoter-associated sequences in LLP1.
[0112] F. Separation of the two chains of P5 using biotin at the 5' end of one of the chains.
[0113] Biotin can specifically combine with Streptavidin or Avidin, so the two chains of P5 are separated after the magnetic beads containing Stre...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com