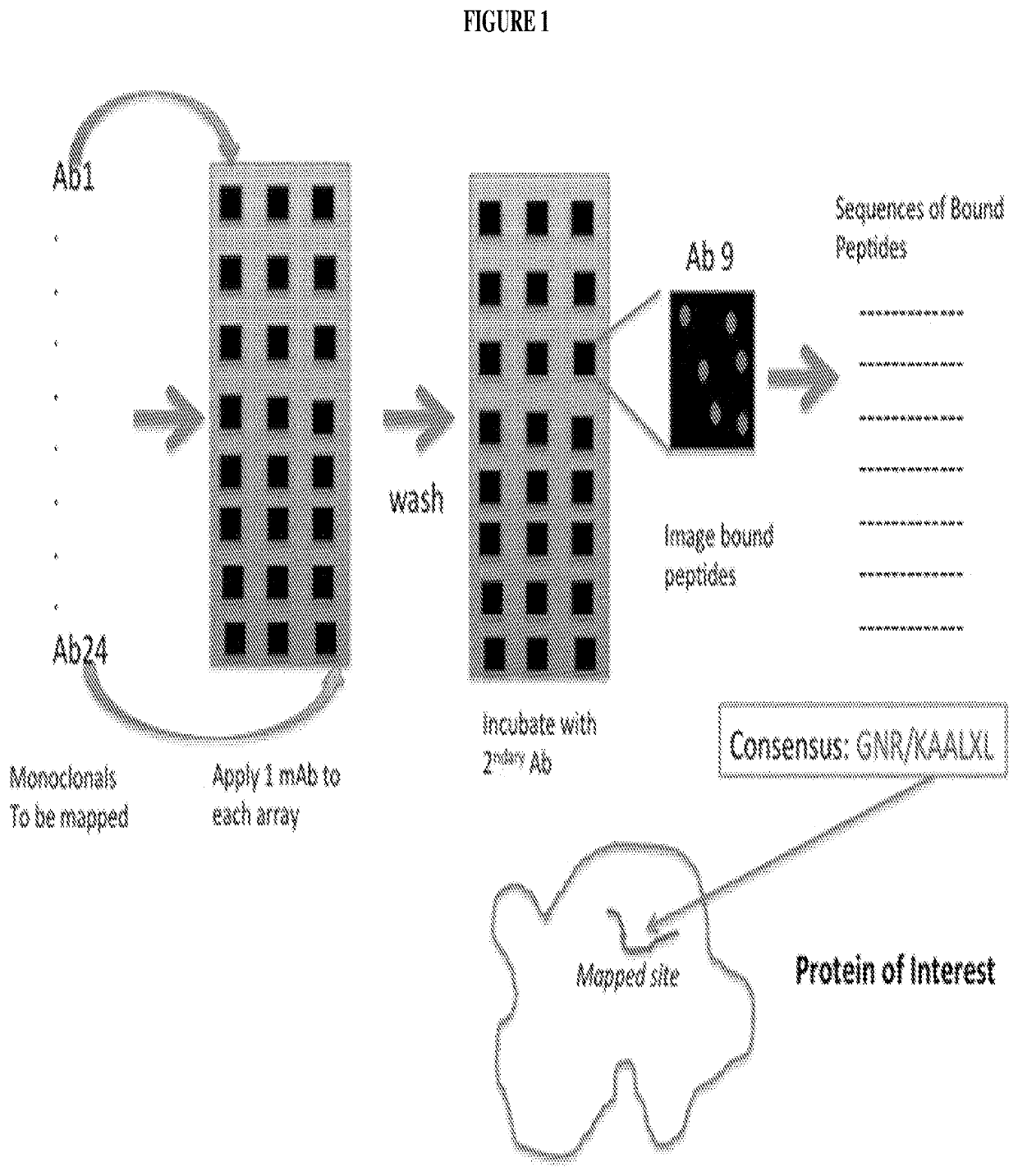
Systems and methods of epitope binning and antibody profiling
a monoclonal antibody and epitope mapping technology, applied in the field of epitope mapping and monoclonal antibody profiling, can solve the problems of requiring costly synthesis or enrichment steps, standard methods developed for mapping antibody epitopes, bacteria, mrna display,
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Epitope Determination in Monoclonal Antibodies
[0077]Experiments were designed to determine whether one could predicatively map epitopes to well-characterized monoclonal antibodies. Eight antibodies with reactivity to a known linear sequence were chosen and analyzed.
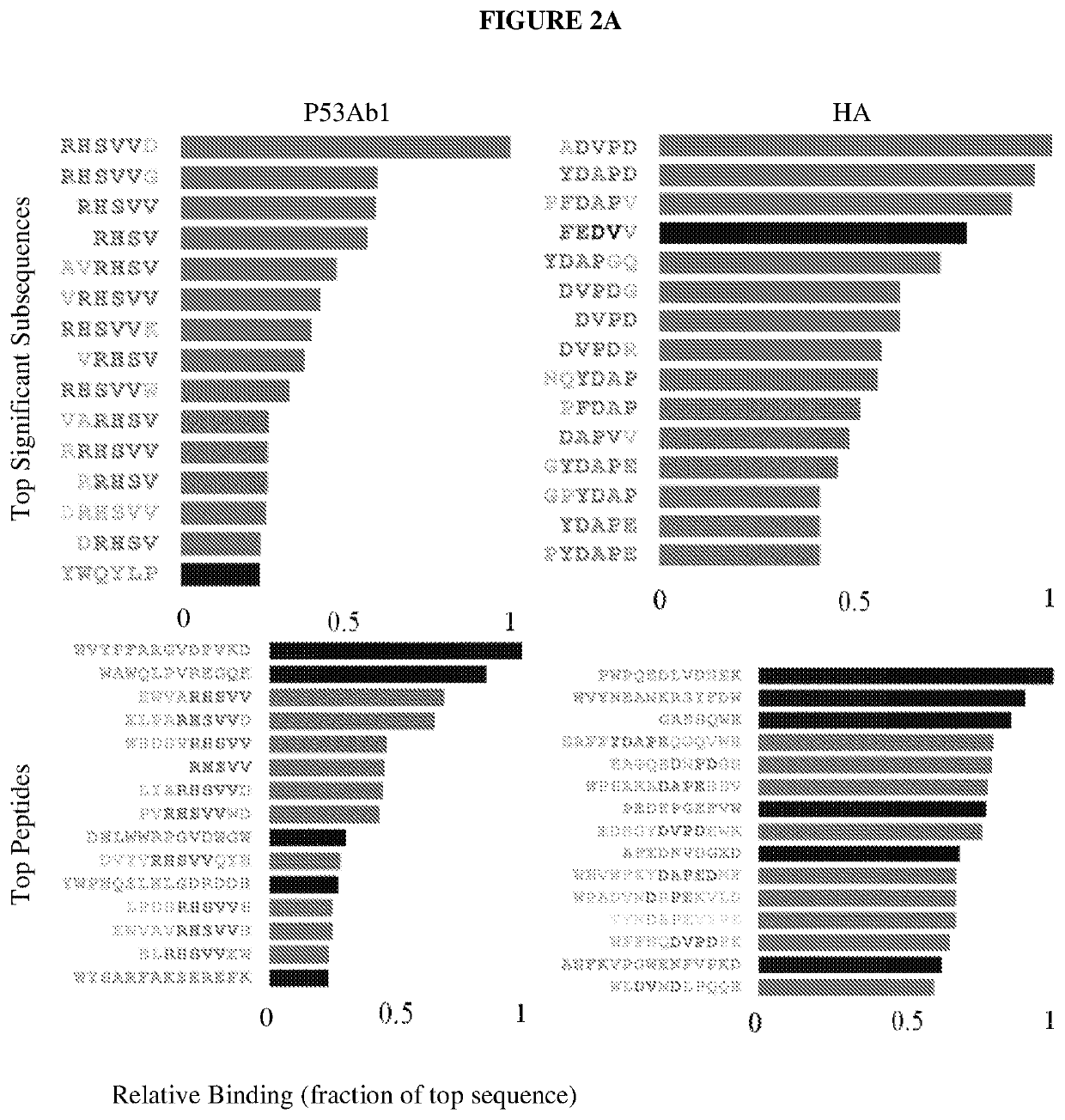
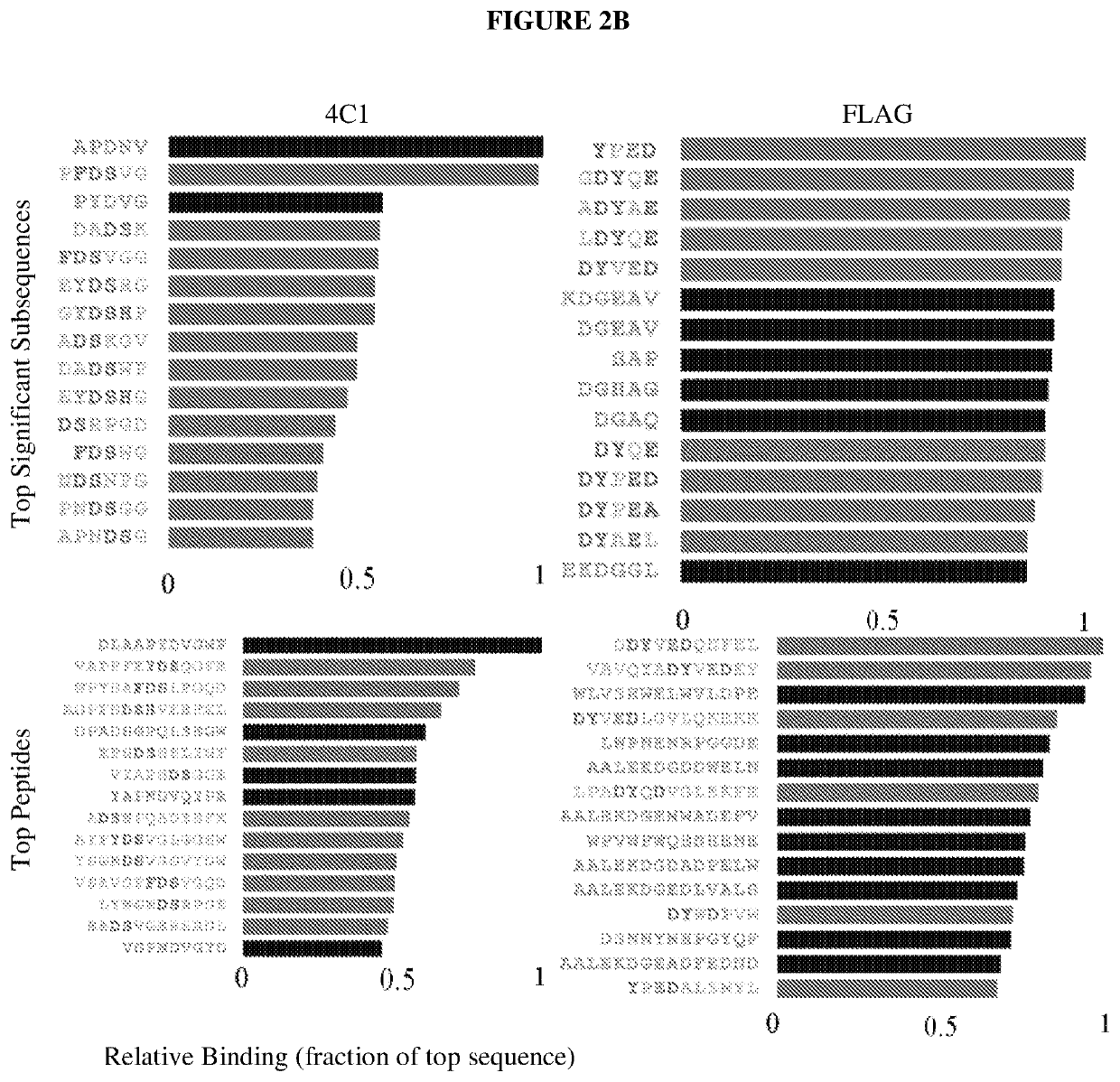
[0078]Table 1 lists peptides and binding intensities for the eight different monoclonal antibodies. Monoclonal antibodies were used to test the motif search analysis algorithm. The highest rated subsequences were related to the true epitope and to each other to an extent that ensured the emergence of a conserved motif with strong association to the epitope sequence. Ab, antibody; GRAVY, grand average of hydropathicity index (Legutki J. B., Nat. Commun. 5:4785, (2014)).
TABLE 1Monoclonal antibodies used in this studyMeanMappedAbsignalpredic-EpitopenameImmunogenIsotypepIGRAVYintensityativelyEEDFRVA10Human Pol IIIgG2b4.1−1.34911NoSDLWKLp53ab8Human p53IgG2b,5.6−0.36243NoIgG2aQAFDSH4C1HumanIgG2a5.1−1.1971YesinsulinreceptorRHSVV...
example 2
Groupwise Epitope Determination in Patient Sera
[0088]Eight cohorts representing seven different diseases and one group of healthy volunteers were tested using the described methods.
[0089]FIG. 5A shows the top 10 most commonly appearing and significant subsequences in serum samples from the indicated disease cohorts. The number of patients within that cohort for which that sequence was called significant is shown in parentheses to the left. The y-axis is categorical and shows each subsequence; the x-axis is the maximum log 10-normalized intensity of the peptide binding on the array for each patient. The total number of samples in each cohort is given as a fraction at the top. Subsequences with exact matches to proteins within the pathogen are indicated with vertical red bars. The top ranked sequences are listed in Table 3 that shows discovered epitope sequences and their proposed antigen mappings as described below (Example 3).
TABLE 3Proposed epitope mappings for disease cohortsKnown...
example 3
Identification of Consensus Sequences in Pathogen Proteomes
[0092]In order to test whether the groupwise consensus motifs (FIGS. 4A-B) corresponded with true epitopes, the Immune Epitope Database was searched for exact substring matches to sequences from these lists. Despite the small size of this database, the sequence AVHAD from dengue was present in the database and indicated as an epitope from the NS1 protein in two dengue strains (E-value: 5×10−4).
[0093]Further analysis of the other cohorts revealed additional matches to antigenic proteins. The sequence EDAK from Borrelia mapped to known antigen OspF (E-value: 4.6), and DYAFG from syphilis mapped to a lipoprotein in several strains of T. pallidum (E-value: 0.27). Malaria contained sequences SNKQG and RLKEP (FIG. 7), both of which mapped to the ring-infect erythrocyte surface antigen (RESA) protein in P. falciparum 3D7 (E-value: 0.072), and another sequence (DAFEY) mapping to one of the pfEMP1 variants in P. falciparum (E-value: ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| length | aaaaa | aaaaa |
| length | aaaaa | aaaaa |
| length | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com