Molecular evidence platform for auditable, continuous optimization of variant interpretation in genetic and genomic testing and analysis
a molecular evidence platform and optimization technology, applied in the field of molecular evidence platforms for auditability and optimization of variant interpretation in genetic and genomic testing and analysis, can solve the problems of 11% classification instability, 11% classification uncertainty, and low accuracy of molecular variant clinical significan
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
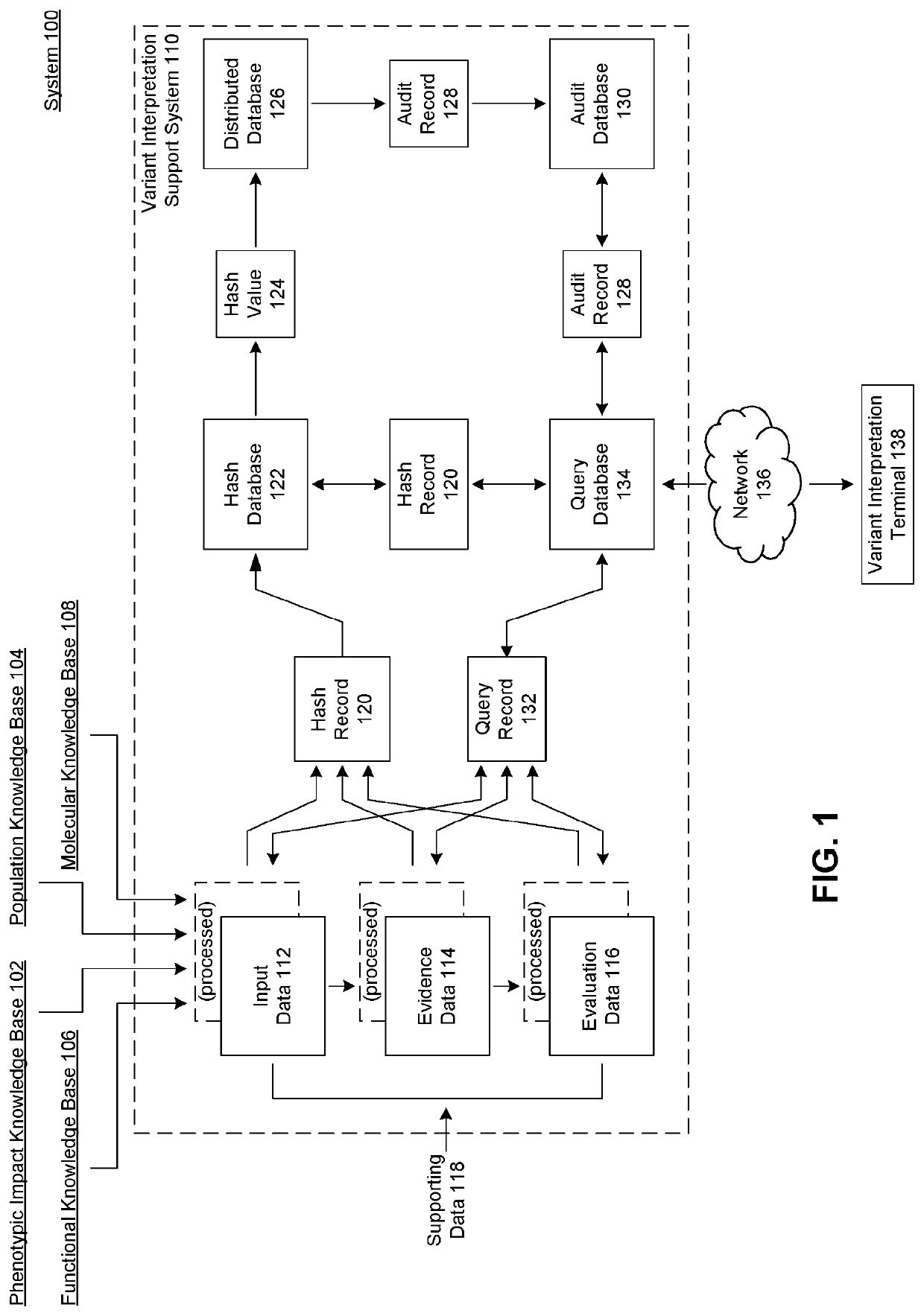
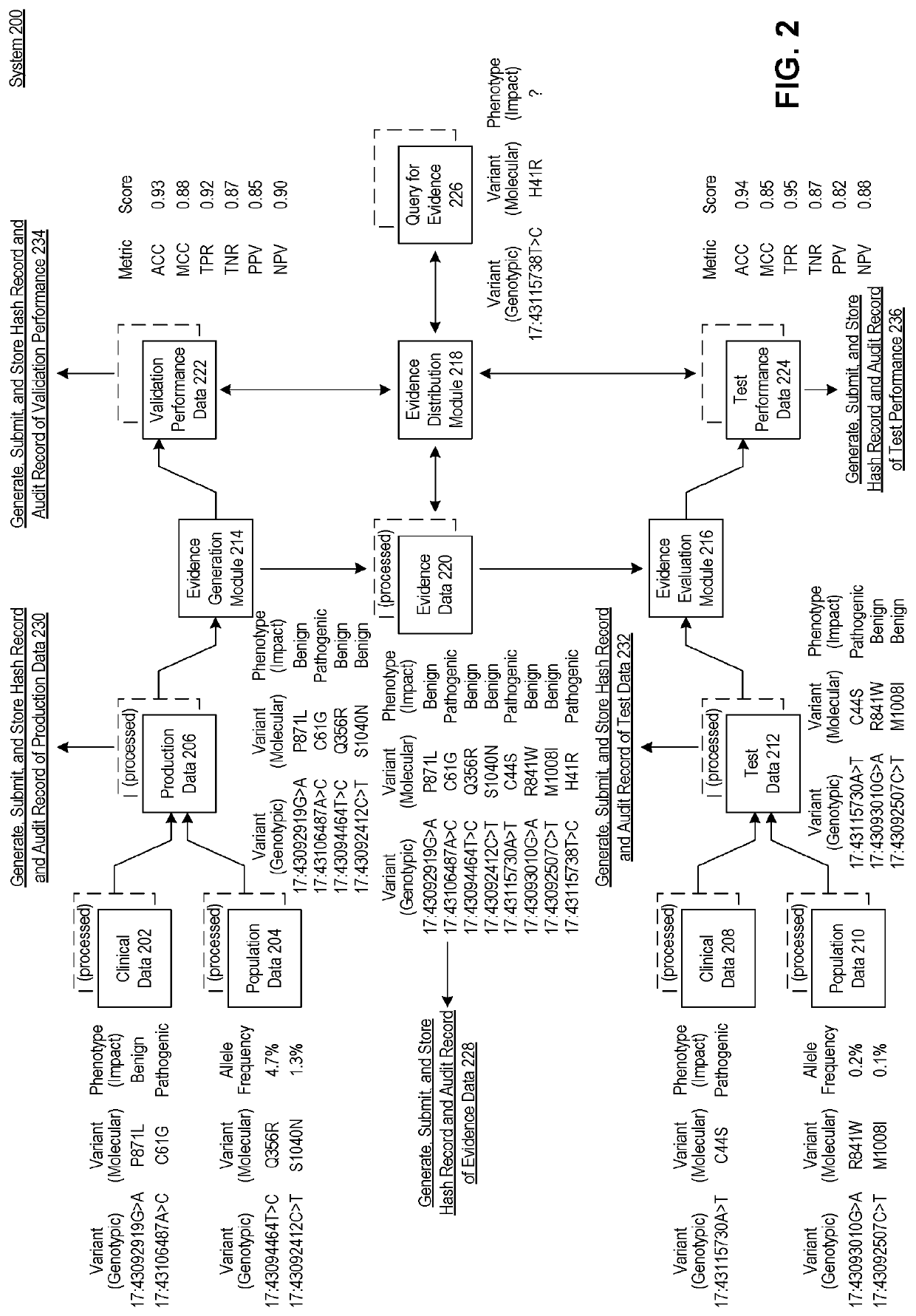
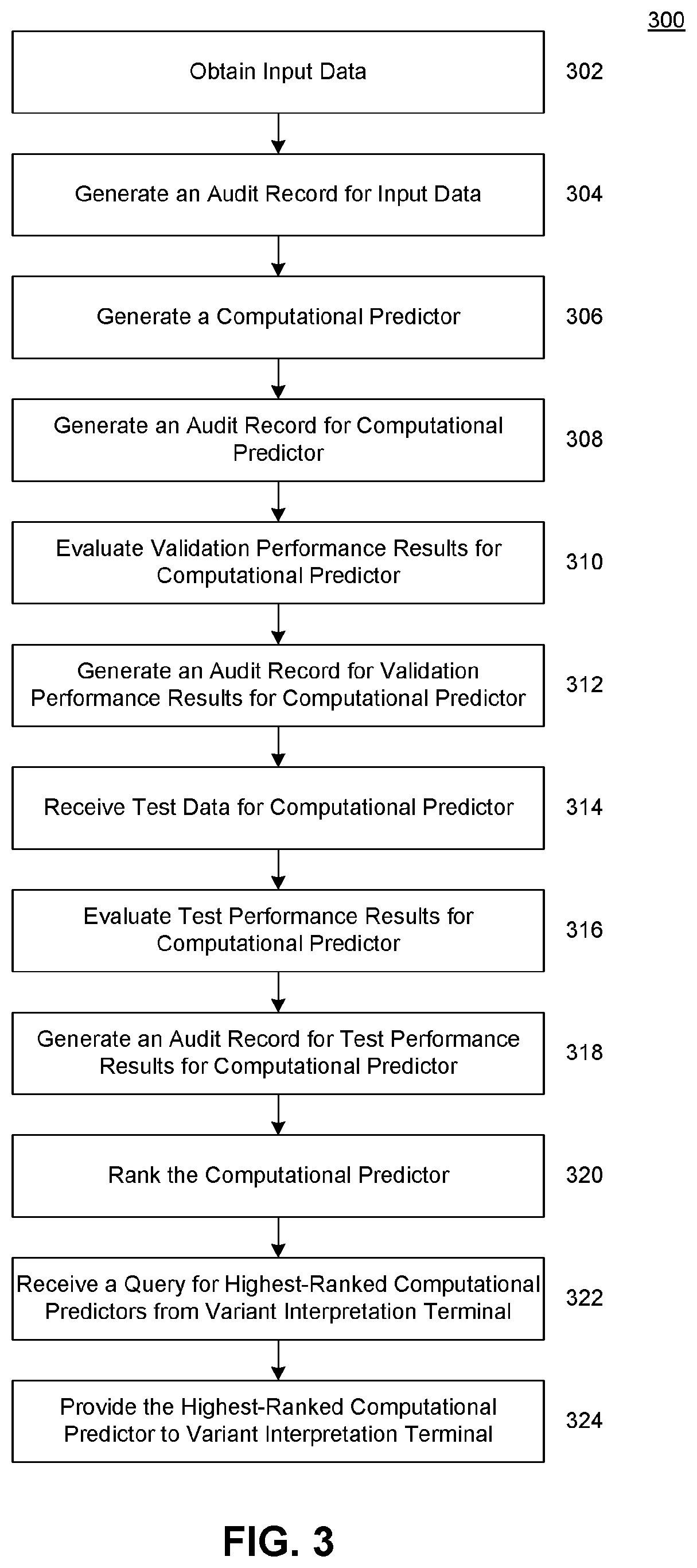
[0028]Provided herein are system, apparatus, device, method and / or computer program product aspects, and / or combinations and sub-combinations thereof, for optimizing the determination of the phenotypic (e.g., clinical or non-clinical) impact (e.g., pathogenicity, functionality, or relative effect) of molecular variants identified in molecular tests, samples, or reports of subjects—such as genotypic (sequence) variants identified in genetic and genomic tests, samples, or reports—by way of regularly incorporating, updating, monitoring, validating, selecting, and auditing the best-performing supporting evidence models for the interpretation of molecular variants across a plurality of evidence classes.
[0029]In some aspects, each evidence model can constitute a system of unique molecular variants and their associated (e.g., clinical or non-clinical) phenotypic impact (e.g., pathogenicity, functionality, or relative effect). As would be appreciated by a person of or ordinary skill in the ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com