Method of nucleic acid cassette assembly
a nucleic acid cassette and assembly technology, applied in the field of molecular biology, can solve the problems that conventional genetic engineering techniques are limited to allowing manipulation of existing sequences, and achieve the effect of complementing or facilitating the ability of the genom
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Mycoplasma Comparative Genomics to Identify Genes in a Minimal Gene Set
[0051]Mycoplasma Comparative Genomics. The 13 complete and 2 partial genome sequences currently in the inventors' dataset comprise an in silico laboratory.
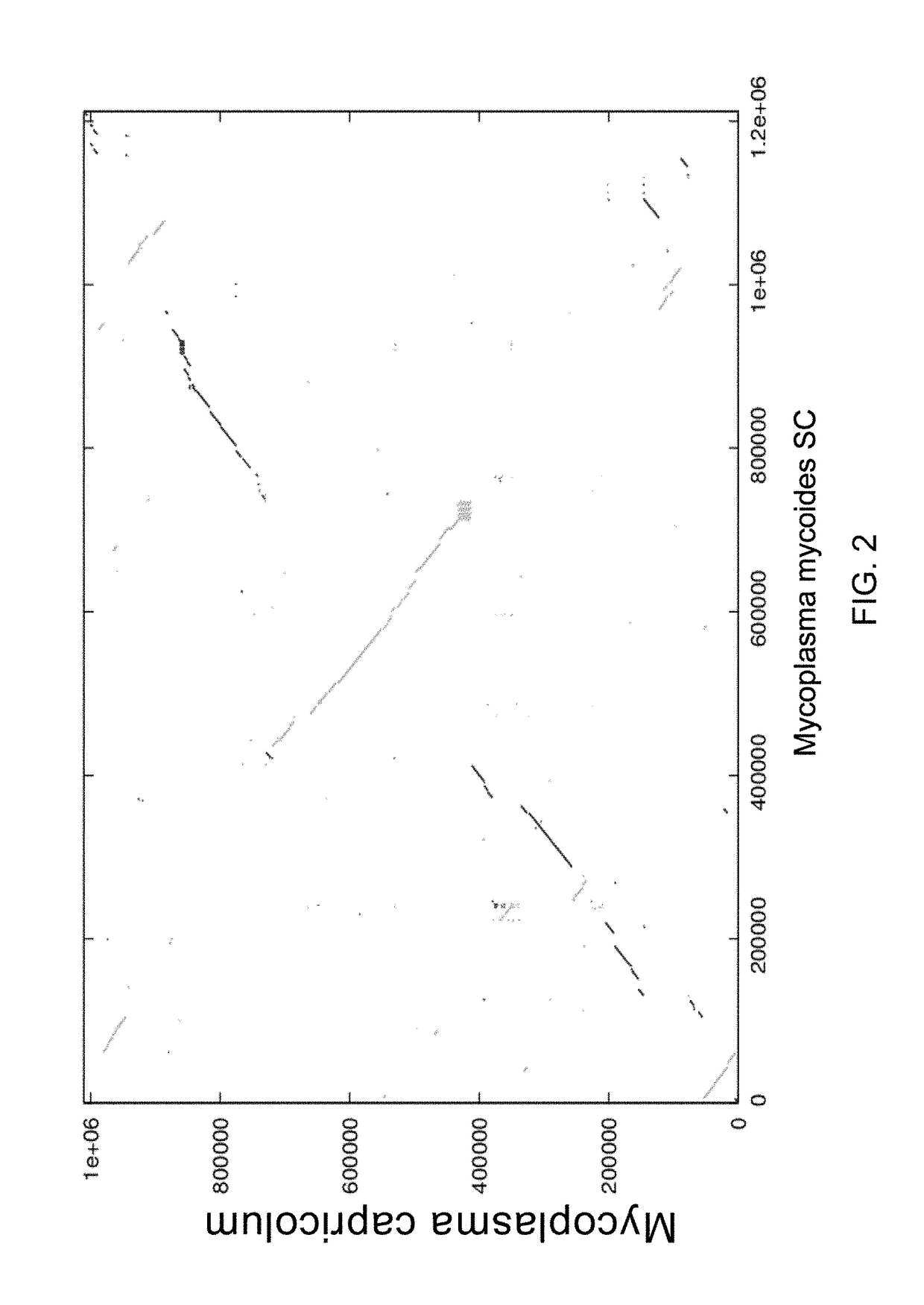
[0052]Comparisons of pairs of mycoplasma genomes using the whole genome alignment tool MUMMER shows between species that are closely related such as M. capricolum, M. mycoides SC, and Mesoplasma florum, genome rearrangements are symmetrical about an axis passing through the origins of replication and points that bisect the genome equally. The direction of transcription of rearranged genes almost always stays the same relative to the origin of replication. This phenomenon has been observed for other species bacteria but perhaps never so strikingly as with M. capricolum and M. mycoides SC (see FIG. 2). These reciprocal crossovers suggest that any major removal of DNA from one side of the genome might need to be matched with a similar deletion from the other side ...
example ii
Synthesis of a Mouse Mitochondrial Genome
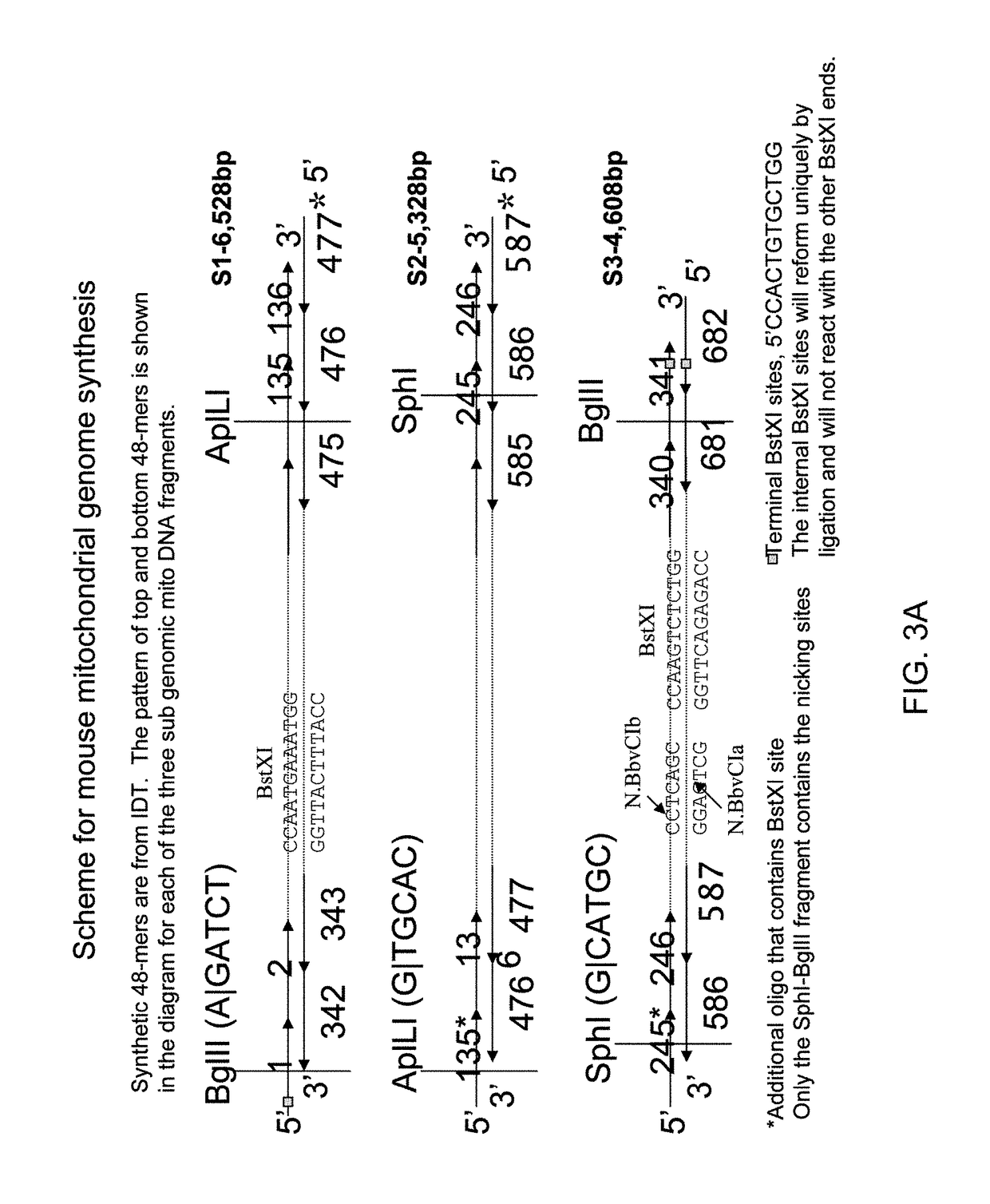
[0056]The mouse mitochondrial genome is a 16,299 bp circular DNA and its sequence has been critically checked. We designed 682 48-mers to assemble it in three overlapping segments; Si (6,528 bp), S2 (5,328 bp), and S3 (4,508 bp) as diagrammed in FIG. 3A.
[0057]We assembled each of these three pieces by the method described in Smith et al. (2003), supra, modified to reduce the heating damage to the synthetic product. The gel shown in FIG. 3B illustrates products from one such modified procedure that dramatically reduces the time spent at high temperature.
[0058]The synthesis of three overlapping segments comprising the entire mouse mitochondrial genome illustrates that combinations of PCA and PCR can routinely assemble 5-6 Kris Prather segments of DNA and validates our plan to build 5 Kris Prather cassettes for the assembly of a cellular genome.
example iii
Synthesis of 5 kb Cassettes of an Essential Region of the M. Genitaliumgenome
[0059]5 kb cassettes are constructed to generate a synthetic copy of an essential region of the M. genitalium genome—the ribosomal protein genes MG149.1 through MG181. This 18.5 kb region is flanked by genes that tolerate transposon insertions (MG149 and MG182). Sets of 386 top strand and 386 bottom strand oligonucleotides, of 48 nt, were synthesized to cover this region. These nucleic acids are illustrated in FIG. 4.
[0060]The assembly of four overlapping segments (cassettes) comprising these oligonucleotides is performed.
[0061]Using these techniques, cassettes of, for example, 4-6 kb can be constructed that include gene sets of interest (e.g., a minimal genome from a unicellular microorganism), and can be “mixed and matched” with, or altered by substitutions from, e.g., other species, to obtain a custom made genome, which can be introduced into a vesicle or ghost cell for testing, as described above. Synt...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com