C-Type Lectin Fold as a Scaffold for Massive Sequence Variation
a lectin and sequence variation technology, applied in the field of binding proteins, can solve the problems of limited variability in the sequence of proteins, immunoglobulins, and amino acid residues with side chains that are not exposed to the exterior solvent, and achieve the effects of stable structure, high tolerance, and stable structur
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Crystal Structures of MTD Variants
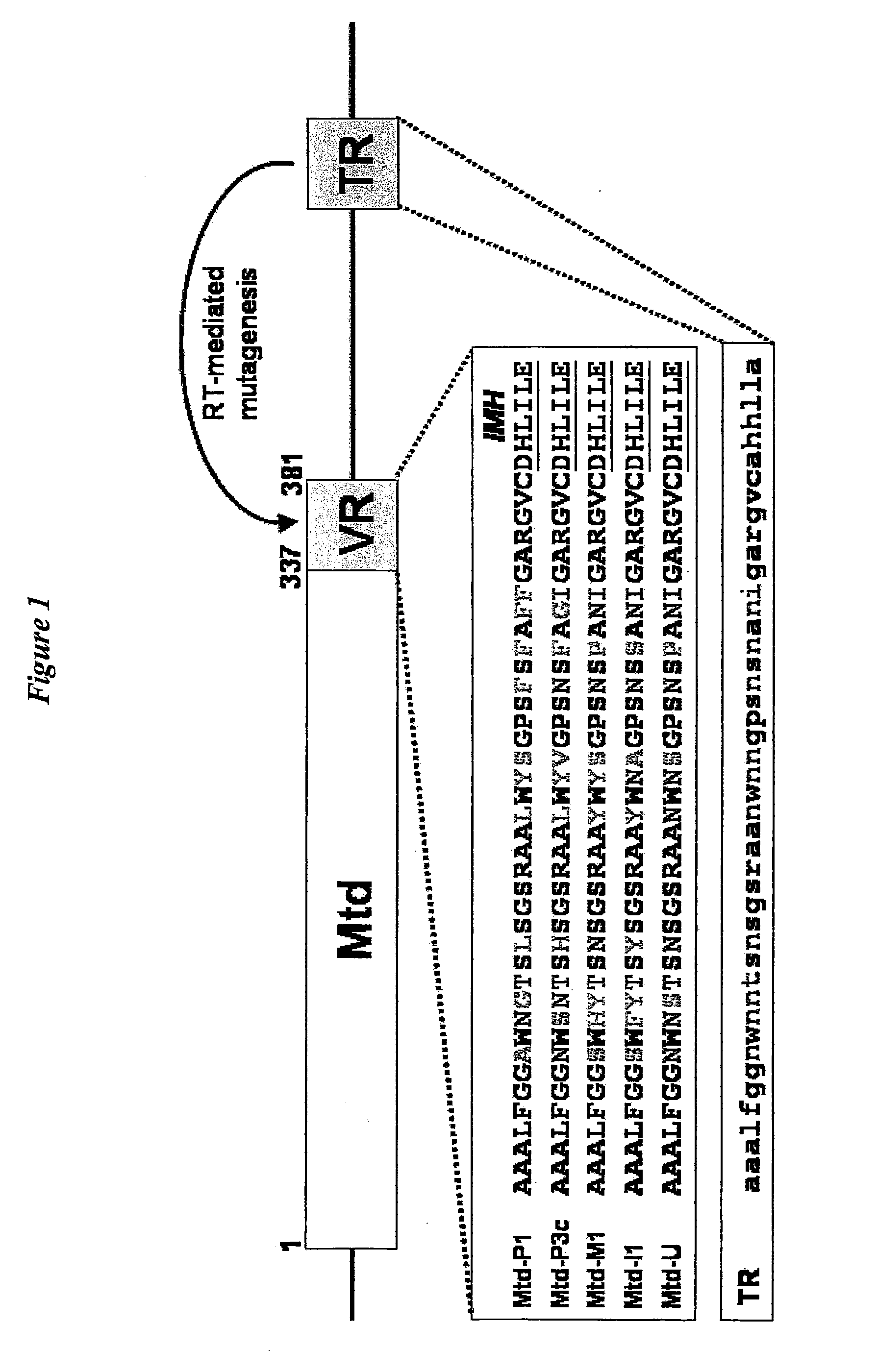
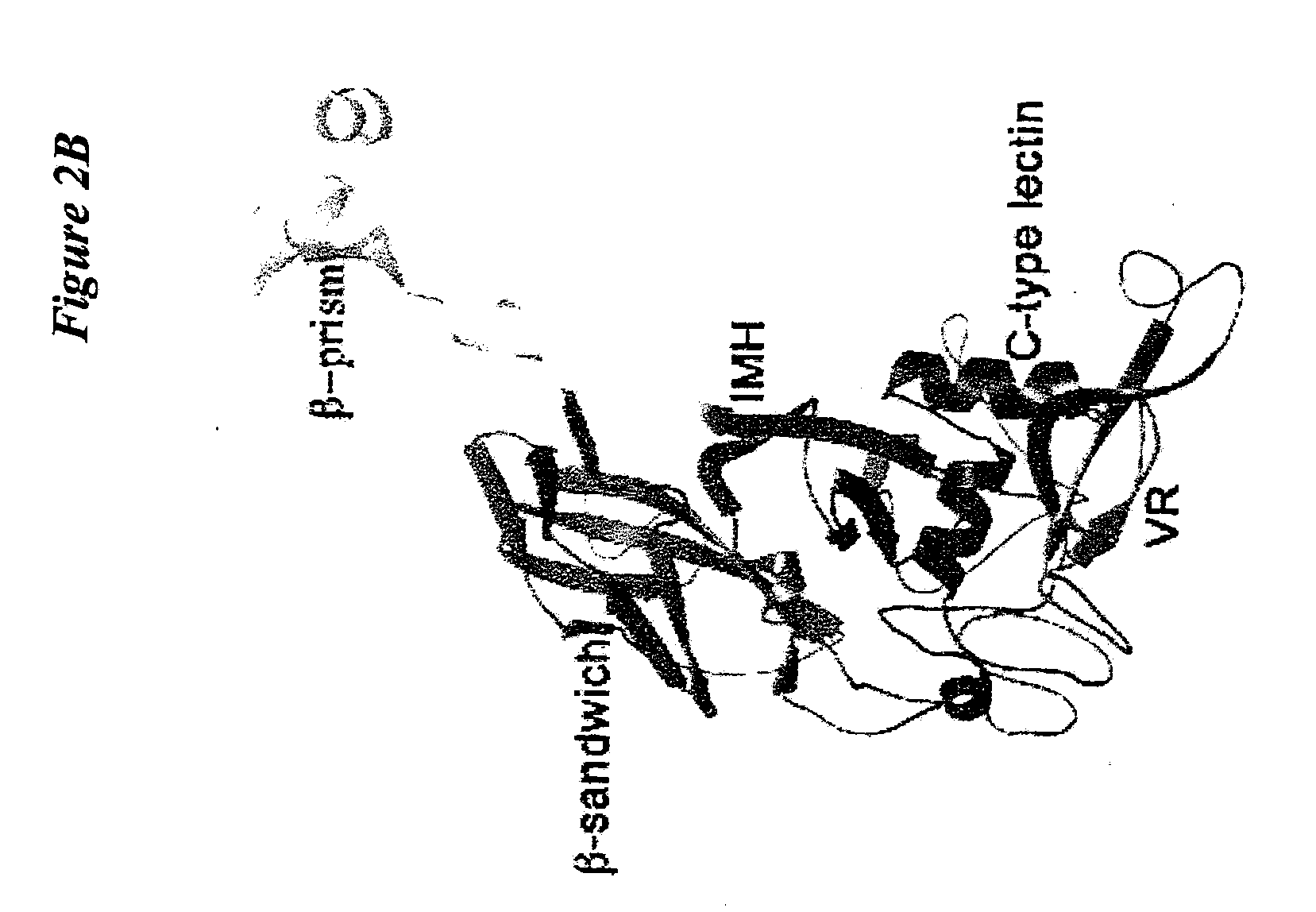
[0147]Structural comparison of Mtd-P1,-3c, -M1, I1, and -N1 were used to discover that the main chain conformation of the CTL domain is remarkably invariant, despite half of the variable residues being on loop regions (FIG. 3C). The binding site in these variants is highly well ordered, having average main chain B-factors ranging from ˜9 Å in Mtd-P1 to −24 Å2 in Mtd-M1 and with density visible for all but one side chain (Phe-346 in Mtd-I1). Providing stabilization to these loops in Mtd are two features unique to the Mtd CTL-fold, namely the two inserts and trimeric assembly.
[0148]The inserts form hydrogen bonds to VR, including three to side chains of three invariant serines in VR. Ser-270 and Glu-267 from the second insert form hydrogen bonds to the invariant VR residues Ser-351 and Ser-353, respectively (FIG. 3D), and main chain atoms of the first insert form hydrogen bonds to invariant VR residue Ser-365 (not depicted). These interactions are sup...
example 2
Basis of MTD to Ligand Interactions
[0150]To understand the basis of Mtd interactions with its ligand, a cell surface receptor, we characterized association between Mtd-P 1 and the Bordetella receptor pertactin. The pertactin ectodomain (Prn-E) was incubated with Mtd variants and found by a coprecipitation assay to associate most strongly with Mtd-P1 but also with Mtd-3c and Mtd-M1. As a measure of specificity, Prn-E was not found to associate with Mtd-I1 or Mtd-N1. The three Mtd variants that are found to bind pertactin have in common the variable residue Tyr-359, previously shown by sequence comparison to be a consistent determinant for pertactin interaction. The presence of a tyrosine residue in the binding pocket is consistent with the presence of a number of hydrophobic surface-exposed patches on Prn-E (see Emsley, P., et al. Structure of Bordetella pertussis virulence factor P.69 pertactin. Nature 381, 90-2 (1996)). The maintenance of Pm affinity in some of these Mtd variants a...
example 3
CTL-Fold in Other DGRs
[0152]A number of other putative DGRs have been identified in phage and bacterial genomes. These resemble the Bordetella phage DGR in having sequence-related reverse transcriptases, similar arrangements of VR and TR, adenines constituting the main differences between VR and TR, and IMH-like elements at the end of VR. However, the putative variable proteins have no obvious sequence relationship to Mtd or other proteins. Because there appears to be no genetic requirement for VR and its IMH element to be positioned at the very C-terminus of a protein, the variations in positioning likely reflects the necessities of protein binding requirements as specified by the CTL-fold. Despite the low sequence identity among these proteins (˜17%), we have been able to use the structure of Mtd along with considerations about variability to construct a sequence alignment consisting of the β2β3β4β4′ sheet of the CTL-fold (see FIG. 5). Most notably, the invariant Mtd binding site ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| length | aaaaa | aaaaa |
| nucleic acid | aaaaa | aaaaa |
| surface | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com