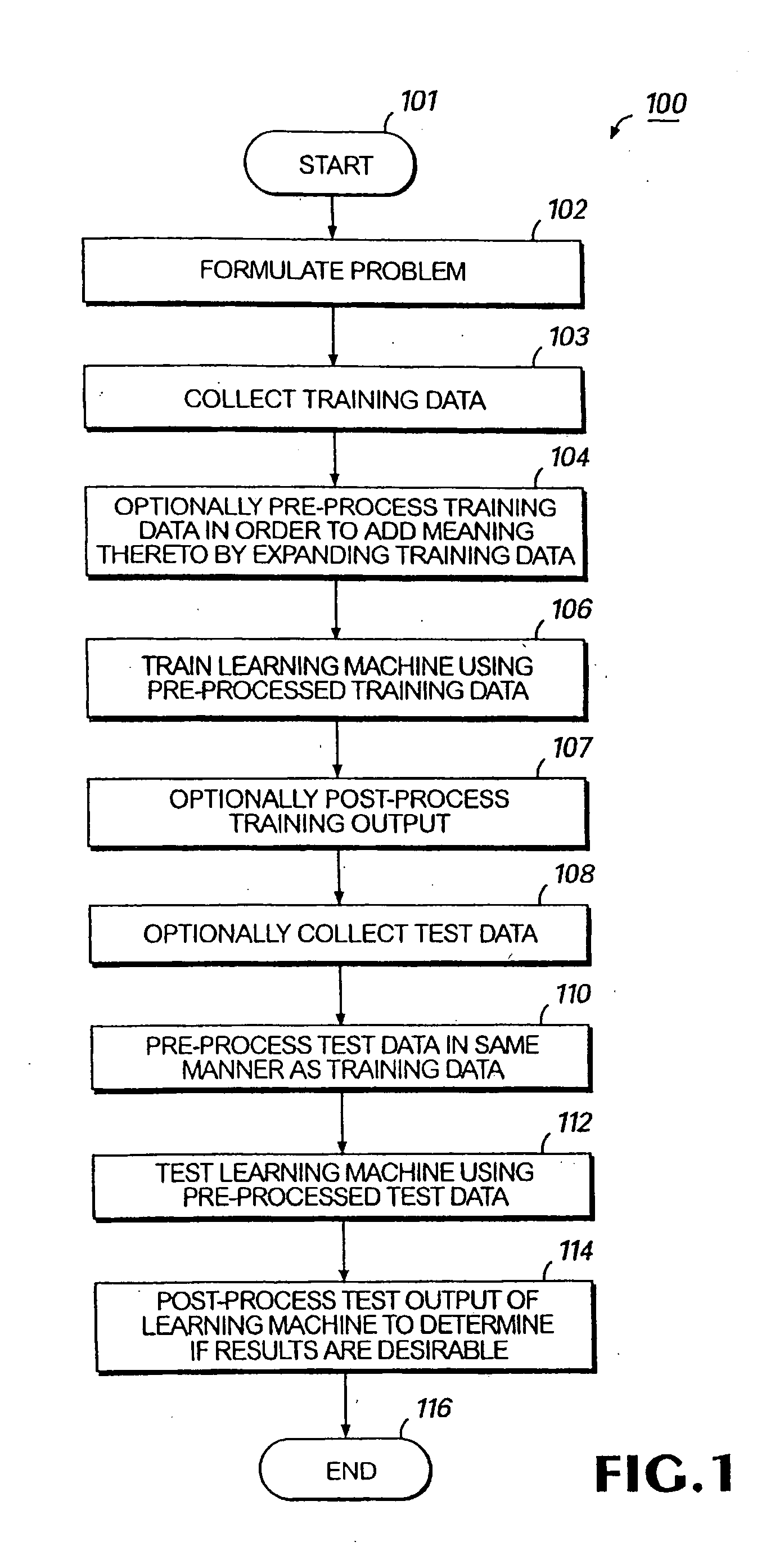
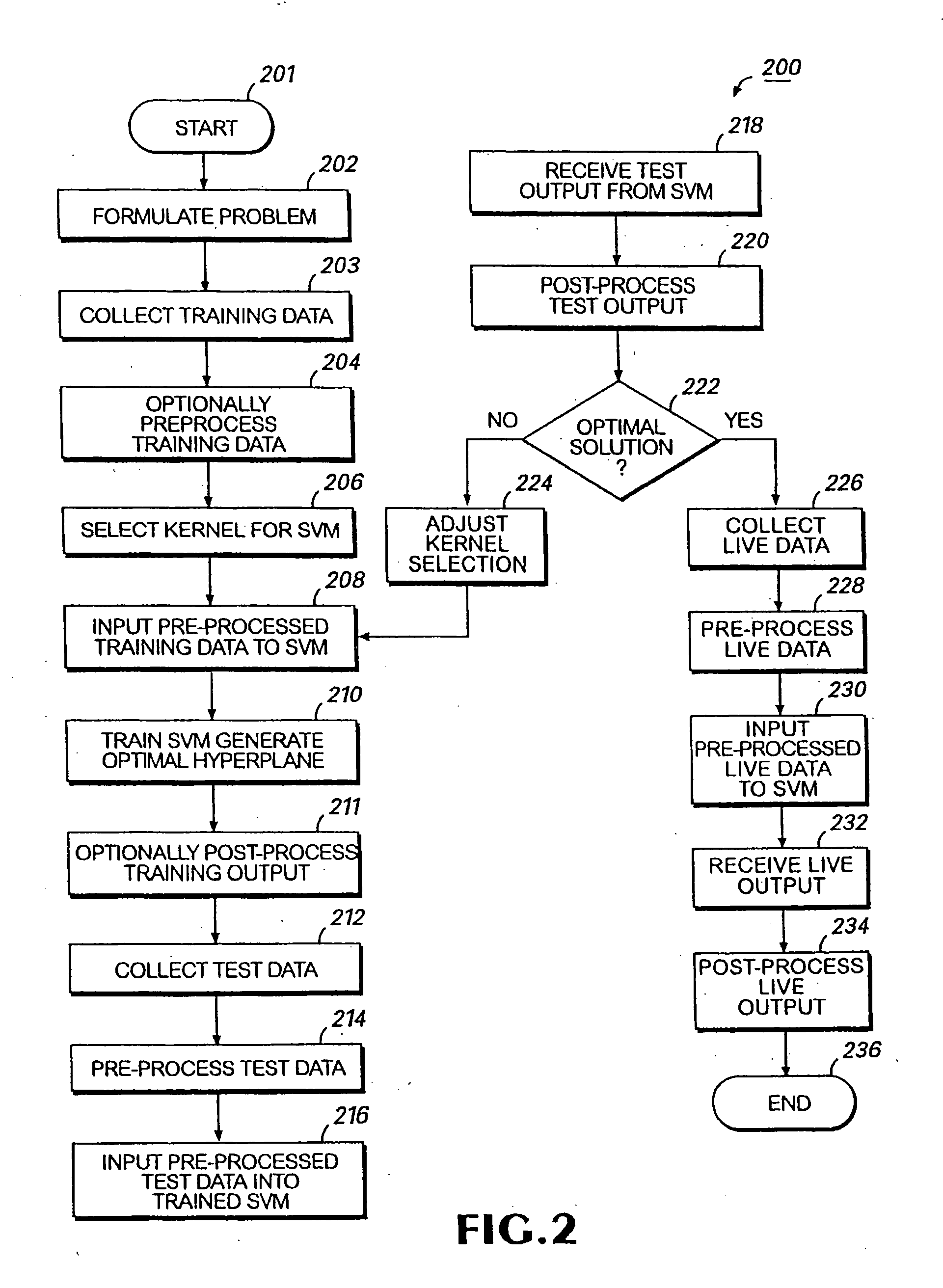
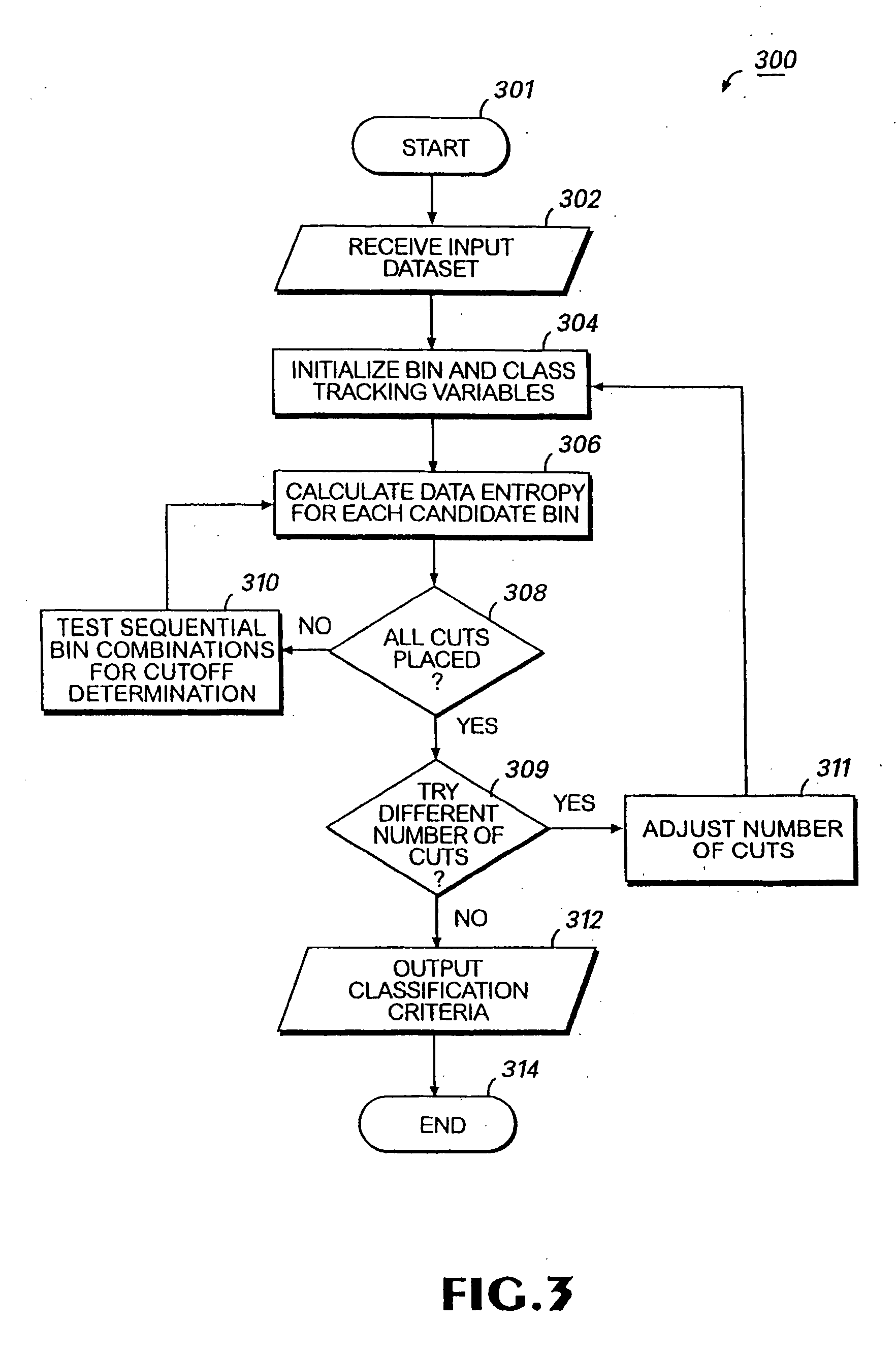
Colon cancer biomarkers
a biomarker and colon cancer technology, applied in the field of colon cancer biomarkers, can solve the problems of impede the derivation of useful information, and require multiple steps to analyze, so as to enhance the ability of learning machines to discover knowledge, and improve the quality of generalizations
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
EXAMPLE 1
Analysis of Gene Patterns Related to Colon Cancer
[0145] Errorless separation can be achieved with any number of genes, from one to many. Preferred methods comprise use of larger numbers of genes. Classical gene selection methods select the genes that individually classify the training data best. These methods include correlation methods and expression ratio methods. They eliminate genes that are useless for discrimination (noise), but do not yield compact gene sets because genes are redundant. Moreover, complementary genes that individually do not separate well the data are missed.
[0146] A simple feature (gene) ranking can be produced by evaluating how well an individual feature contributes to the separation (e.g. cancer vs. normal). Various correlation coefficients are used as ranking criteria. The coefficient used is defined as:
P=(μ1−μ2) / (σ1+σ2)
where μi and σi are the mean and standard deviation of the gene expression values of a particular gene for all the patients...
example 2
EXAMPLE 2
Leukemia Gene Discovery
[0246] The data set, which consisted of a matrix of gene expression vectors obtained from DNA microarrays, was obtained from cancer patients with two different types of leukemia. The data set was easy to separate. After preprocessing, it was possible to find a weighted sum of a set of only a few genes that separated without error the entire data set, thus the data set was linearly separable. Although the separation of the data was easy, the problems present several features of difficulty, including small sample sizes and data differently distributed between training and test set.
[0247] In Golub, 1999, the authors present methods for analyzing gene expression data obtained from DNA micro-arrays in order to classify types of cancer. The problem with the leukemia data was the distinction between two variants of leukemia (ALL and AML). The data is split into two subsets: A training set, used to select genes and adjust the weights of the classifiers, an...
PUM
| Property | Measurement | Unit |
|---|---|---|
| clock time | aaaaa | aaaaa |
| clock time | aaaaa | aaaaa |
| mass spectrophotometry | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com