Methods and devices for measuring differential gene expression
a technology of differential gene expression and measurement method, which is applied in the direction of instruments, biochemistry apparatus and processes, material analysis, etc., can solve the problems of inconvenient use, inconvenient use, and inability to identify all sequences in a complex mixture, and achieve the effect of accurate and efficient measuremen
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
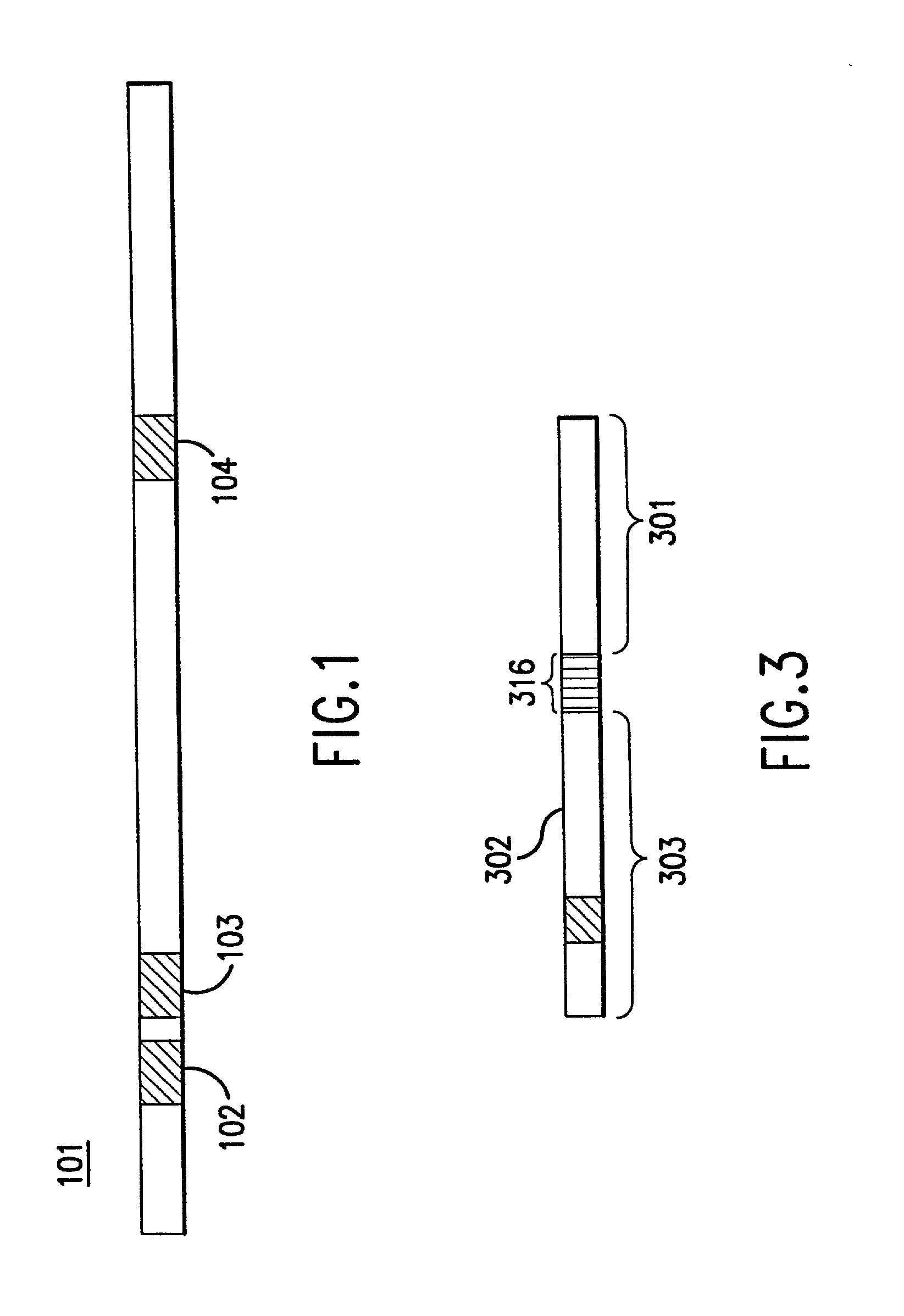
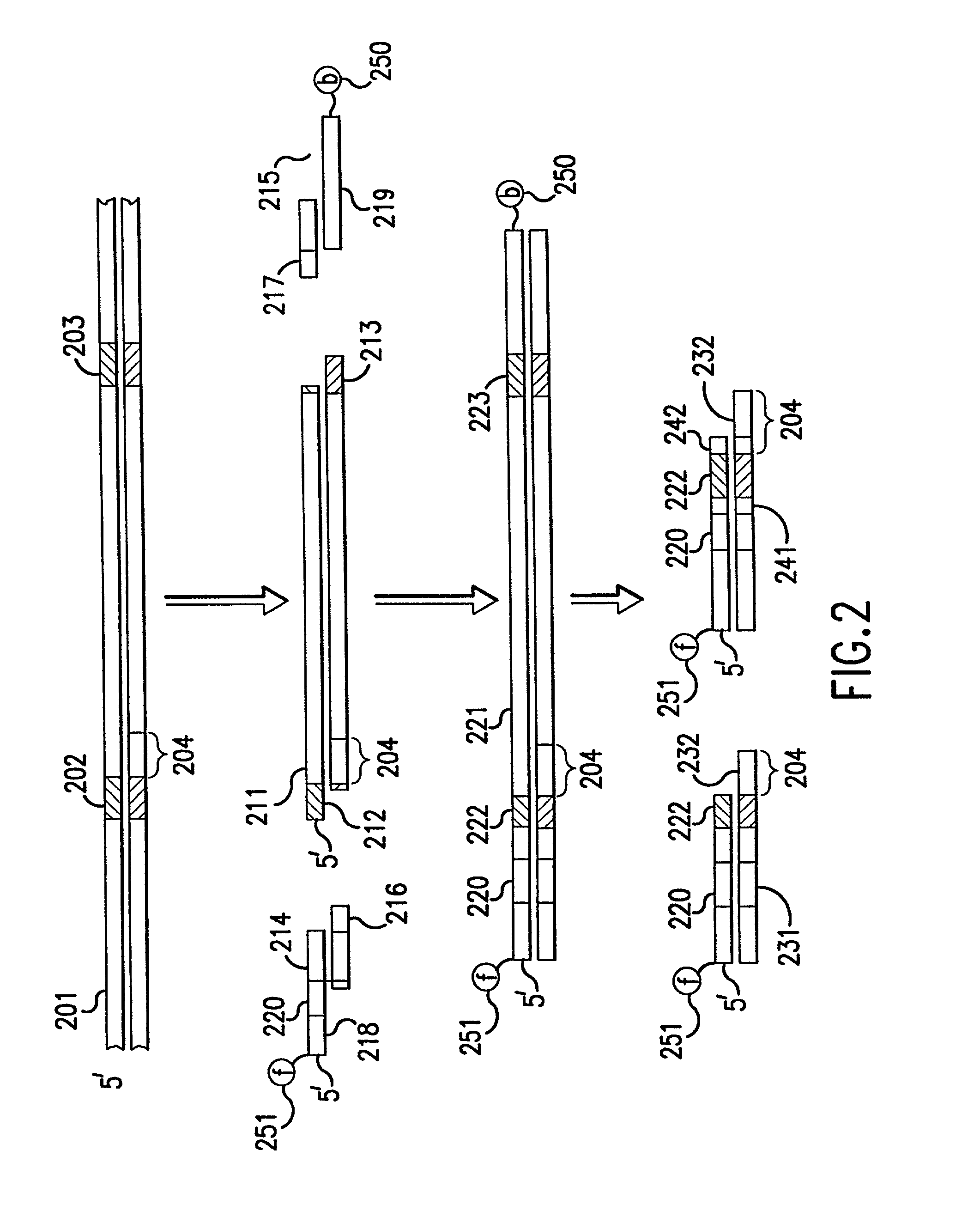
[0064] The instant invention relates to methods and devices for identifying and quantifying nucleic acids in a sample of nucleic acids (also referred to herein as "gene-calling"), and in particular to methods and devices for genomic analysis. Accordingly, this invention can be applied to analysis of gene expression by identifying and quantifying complementary DNA ("cDNA") and to genetic analysis by identifying and quantifying genomic DNA ("gDNA").
[0065] The gene-calling methods of this invention start, in general, with a possibly complex sample of nucleic acids, preferably DNAs, and observe the presence of sets of nucleotide subsequences ("subsequence sets") in the nucleic acids of the sample. Then with reference to a database of nucleotide sequences of nucleic acids that may be in the original sample, the methods determine which sequences have the observed subsequence sets, and which observed subsequence sets are not present in any sequence in the database. Accordingly, nucleic aci...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Tm | aaaaa | aaaaa |
| temperature | aaaaa | aaaaa |
| temperature | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com