Method for isolating subpopulations of proteins that engage in protein-protein interactions
A protein separation, protein technology, applied in the field of protein-protein interaction research
Inactive Publication Date: 2005-08-24
BLANCHETTE ROCKEFELLER NEUROSCI INST
View PDF1 Cites 0 Cited by
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Therefore, native protein interactions or interactions resulting from signaling related to physiological processes such as learning or Alzheimer's disease cannot be studied with this technique
Method used
the structure of the environmentally friendly knitted fabric provided by the present invention; figure 2 Flow chart of the yarn wrapping machine for environmentally friendly knitted fabrics and storage devices; image 3 Is the parameter map of the yarn covering machine
View moreImage
Smart Image Click on the blue labels to locate them in the text.
Smart ImageViewing Examples
Examples
Experimental program
Comparison scheme
Effect test
Embodiment Construction
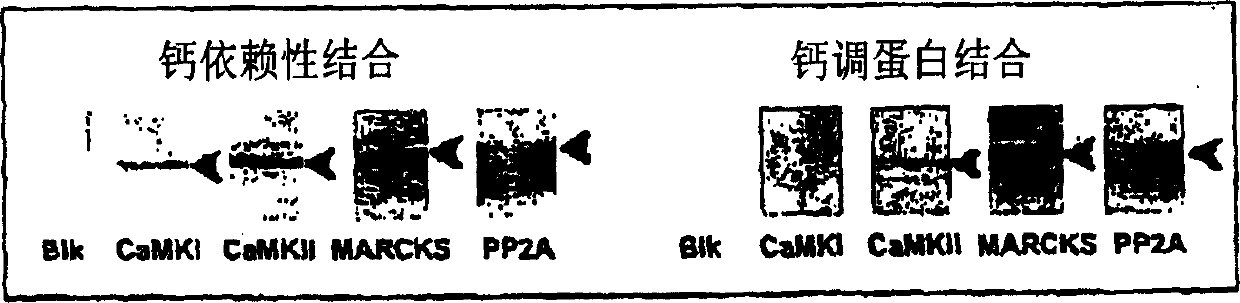
[0031] abbreviation :
[0032] CHAPS: 3-[(3-cholamidopropyl)-diethylamine]-propanesulfonic acid
[0033] EGTA: Ethylene glycol bis(2-aminoethyl ether)-N,N,N',N'-tetraacetic acid
[0034] MARCKS: myristoylation of alanine-rich C kinase substrates
the structure of the environmentally friendly knitted fabric provided by the present invention; figure 2 Flow chart of the yarn wrapping machine for environmentally friendly knitted fabrics and storage devices; image 3 Is the parameter map of the yarn covering machine
Login to View More PUM
 Login to View More
Login to View More Abstract
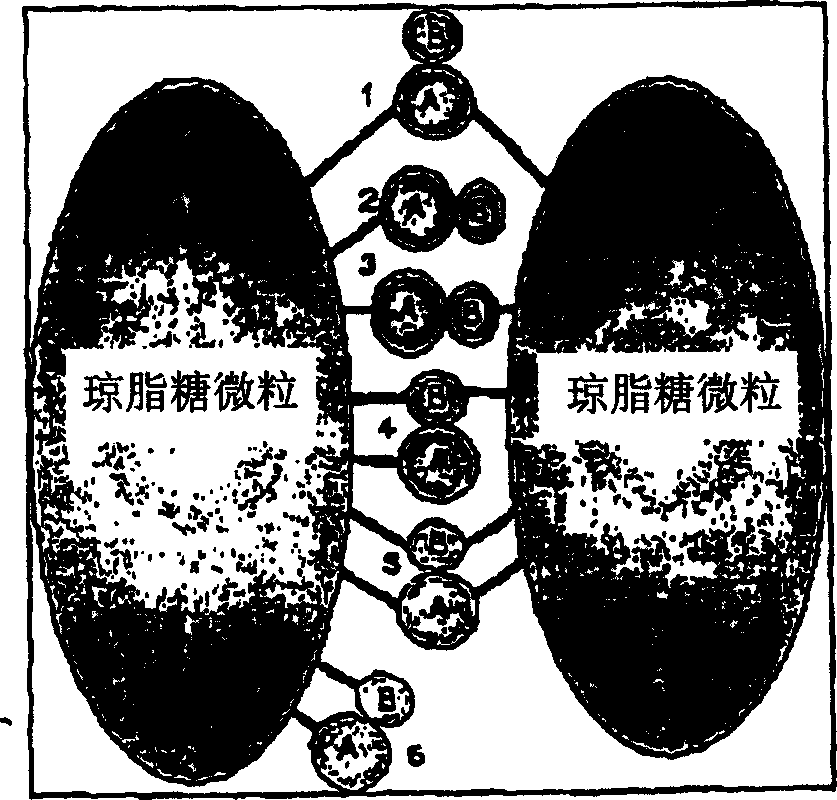
The invention provides a method for isolating and identifying proteins participating in protein-protein interactions in a complex mixture. The method uses a chemically reactive supporting matrix to isolate proteins that in turn non-covalently bind other proteins. The supporting matrix is isolated, and the non-covalently bound proteins are subsequently released for analysis. Because the proteins are accessible to chemical manipulation at both the binding and release steps, identification of the non-covalently bound proteins yields information on specific classes of interacting proteins, such as calcium-dependent or substrate-dependent protein interactions. This permits selection of a subpopulation of proteins from a complex mixture on the basis of specified interaction criteria. The method has the advantage of screening the entire proteome simultaneously, unlike two-hybrid systems or phage display methods which can only detect proteins binding to a single bait protein at a time. The method is applicable to the study of protein-protein interactions in biopsy and autopsy specimens, to the study of protein-protein interactions in the presence of signalling molecules, pharmacological agents or toxins, and for comparison of diseased and normal tissues or cancerous and untransformed cells.
Description
field of invention [0001] The invention relates to the study of protein-protein interactions and is expected to find application in the fields of biochemical signal transduction, proteomics, drug discovery, toxicology and diagnostics. Background of the invention [0002] Protein-protein interactions are fundamental to a variety of physiological processes. Cellular processes such as neuronal signaling, cell development, growth and replication all depend on complex networks of protein-protein and protein-small molecule interactions in the cell. These interactions can be classified as constitutive interactions such as between hemoglobin subunits and signal-dependent interactions such as between cAMP-dependent protein kinase subunits or GTP-binding protein subunits. The complexity of the task of studying these interactions is evident from the potential number of protein interactions: a full screen of 15,000 proteins for binary interactions requires te...
Claims
the structure of the environmentally friendly knitted fabric provided by the present invention; figure 2 Flow chart of the yarn wrapping machine for environmentally friendly knitted fabrics and storage devices; image 3 Is the parameter map of the yarn covering machine
Login to View More Application Information
Patent Timeline
 Login to View More
Login to View More IPC IPC(8): G01N33/53A61K38/00C07K1/00C07K1/14C07K14/00C07K14/47C07K17/00C07K17/06C40B30/04G01N33/543G01N33/566G01N33/68
CPCC40B30/04G01N33/6845G01N33/6842C07K1/14
Inventor D·L·阿尔康T·J·尼尔森
Owner BLANCHETTE ROCKEFELLER NEUROSCI INST
Who we serve
- R&D Engineer
- R&D Manager
- IP Professional
Why Patsnap Eureka
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com