Gene expression amount comparing analysis method
A technology of gene expression and analysis method, which is applied in the field of simultaneous determination of gene expression from various sources by bioluminescence method, can solve the problem that gene expression cannot be directly compared, and achieves the effects of low price, convenient operation and high sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0032] Example 1, the comparison of gene expression in two individuals:
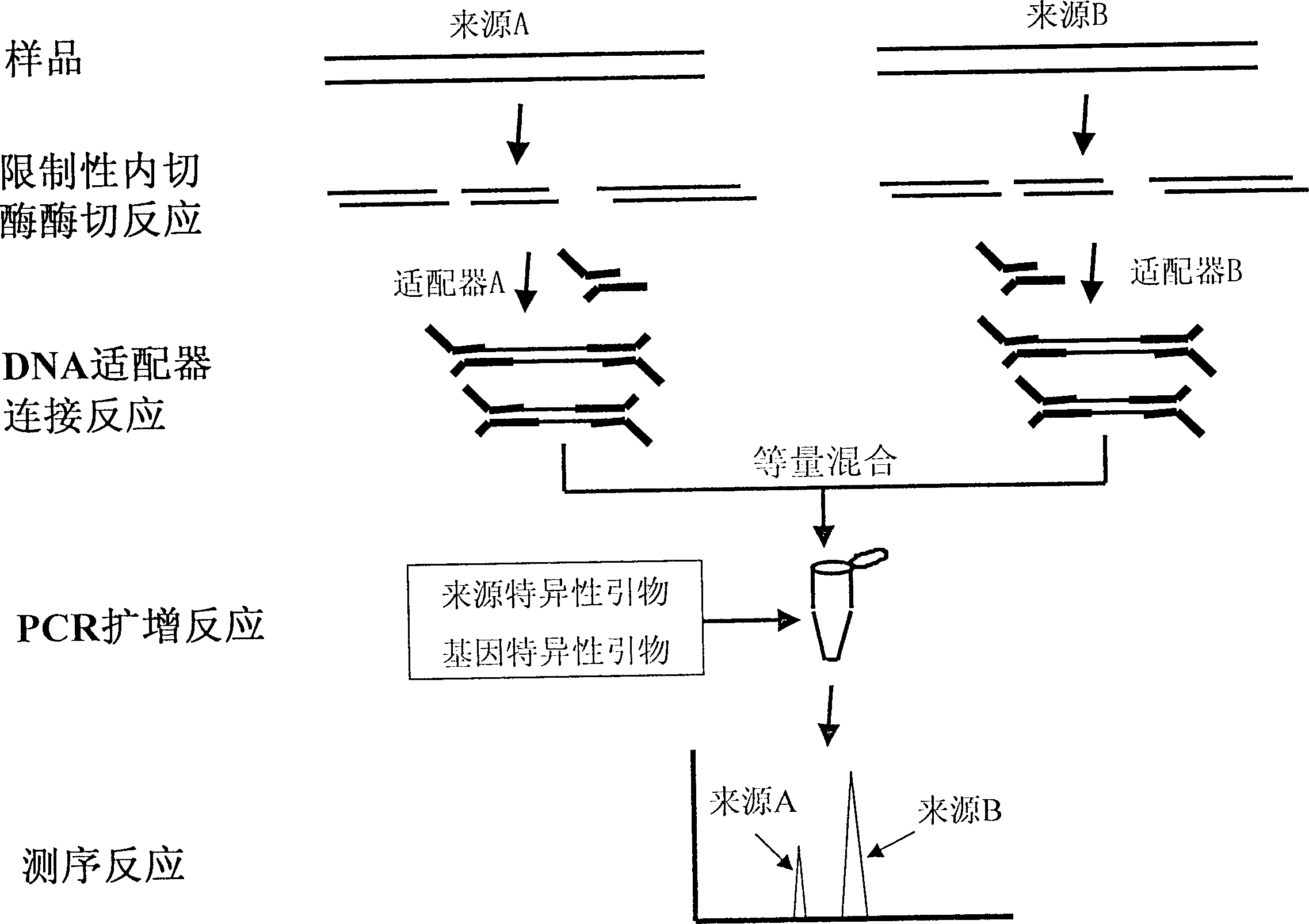
[0033] This example describes the method of cutting cDNA fragments with restriction endonucleases and connecting them with DNA adapters, and then mixing equal amounts of cDNA from source-A and source-B for PCR amplification. The determination process is as follows: figure 2 shown. Taking the human P53 gene as an example, the mRNA was extracted using Gibico TRI20LLS-Reagent TM Standard method of the kit.
[0034] 1. Preparation of cDNA samples (cDNA samples from each source were prepared separately)
[0035] Take 1 μg source-A or source-B mRNA sample, add 0.5 μg Oligo(dT) 12.18 , 5.5 μl H 2 O, and mixed evenly, cDNA first-strand synthesis kit (Gibico Super Script TM Preamplification System for First Strand cDNA Synthesis), after incubation at 0°C for 10 min, add the pre-prepared mixture (4 μl of 5-fold buffer (I), 2 μl of 0.1M dithiothreitol, 1 μl of 10 mM dNTPs, 2 μl h 2 O and 1 μl of 200 U / μl r...
Embodiment 2
[0054] Example 2, using a 96-well reaction plate to simultaneously measure the expression levels of 96 different genes:
[0055] In this embodiment, a common 96-well reaction plate is mainly used to simultaneously measure the expression levels of 96 different genes between the disease group and the healthy group.
[0056] 1. Preparation of Assay Samples
[0057]According to the method of Example 1, extract the mRNA from source A and source B, and make double-stranded cDNA, then cut with restriction enzyme MobI, then add two kinds of DNA adapters representing source A and B respectively, and connect them with ligase Finally, in each well of the 96-well reaction plate, add 1 to 5 μl of the above-mentioned solution as the substrate for PCR amplification, and finally add MP-1, MP-2 and the corresponding GSP primers of each gene for PCR amplification. Amplification reaction, the 5' end of the GSP primer is modified with biotin, and the single-stranded DNA is prepared by the same m...
Embodiment 3
[0061] Example 3, the comparison of gene expression levels from six different sources:
[0062] Usually, microarray chips can only measure the expression of genes from two different sources. If one source is added, a different fluorescent marker needs to be added, which increases the cost of the assay, and usually does not use more than four excitations at the same time. Fluorescent markers with different wavelengths. Since the present invention uses a sequencing method to measure gene expression levels from different sources, and the determined base sequences represent different gene sources, increasing sample sources will not increase any measurement cost.
[0063] Since dATP is an analog of ATP, the background is high, which seriously interferes with the determination. Although it can be replaced by dATPαs, an analog of dATP, the cost of the assay increases. The present invention avoids the use of dATP. When using this method to compare the expression levels of genes from...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com