RNA-protein binding site prediction method and system based on self-attention mechanism
A technology combining sites and prediction methods, applied in the field of bioinformatics, can solve problems such as the need to further improve the prediction accuracy and the insufficient characteristics of the extracted RNA data, so as to improve the prediction accuracy and reduce the experiment time and financial loss.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0071] The present invention will be described in further detail below in conjunction with the accompanying drawings and specific embodiments.
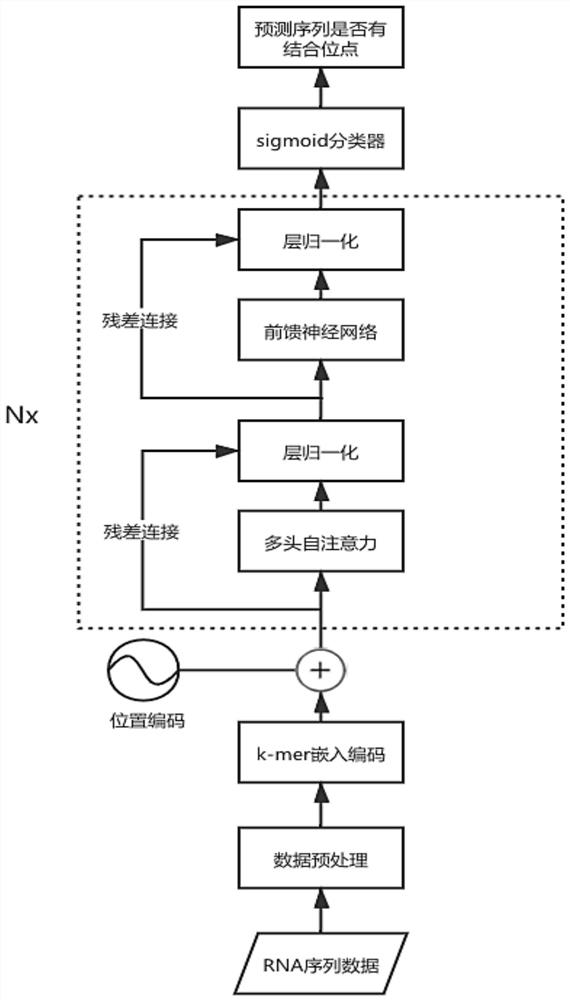
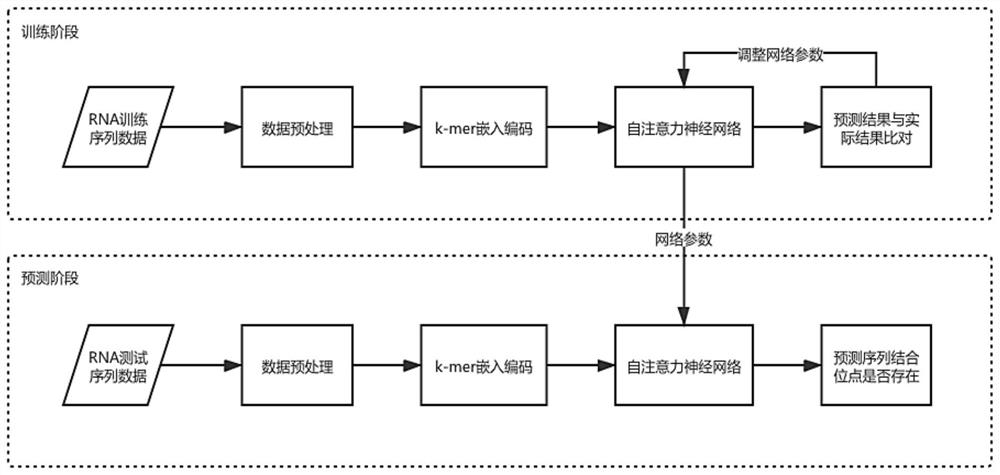
[0072] The present invention proposes a method and system for predicting RNA-protein binding sites based on a self-attention mechanism. This method encodes the contextual relationship between adjacent nucleotides through k-mer embedded coding, and introduces a self-attention mechanism to build a prediction model, giving higher weights to key subsequences so that the network can fully learn key features, thereby improving the model's accuracy. prediction accuracy.
[0073] This embodiment provides a method for predicting RNA-protein binding sites based on the self-attention mechanism, refer to figure 1 and figure 2 , the process is implemented based on python3.8.6-tensorflow2.4.0. The method includes:
[0074] S1: Data acquisition and preprocessing, acquiring RNA sequence data and performing data preprocessing;
[0075] S2: Based...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com