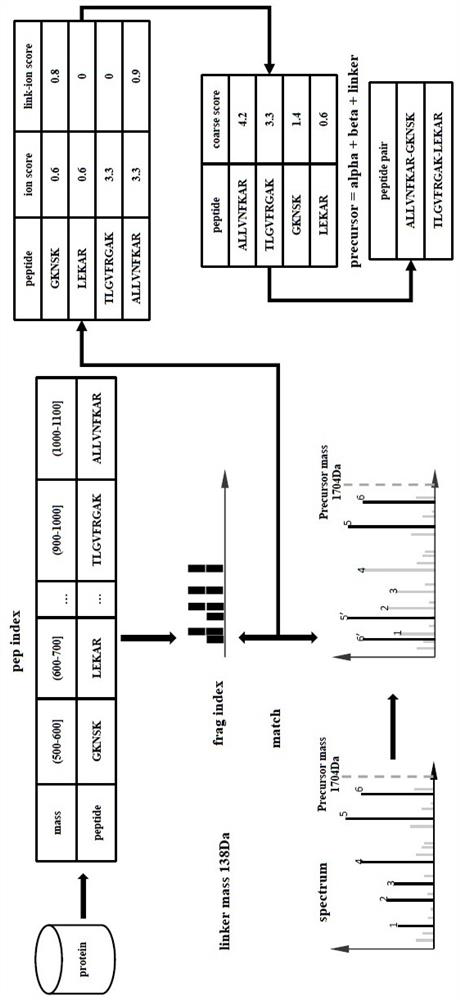
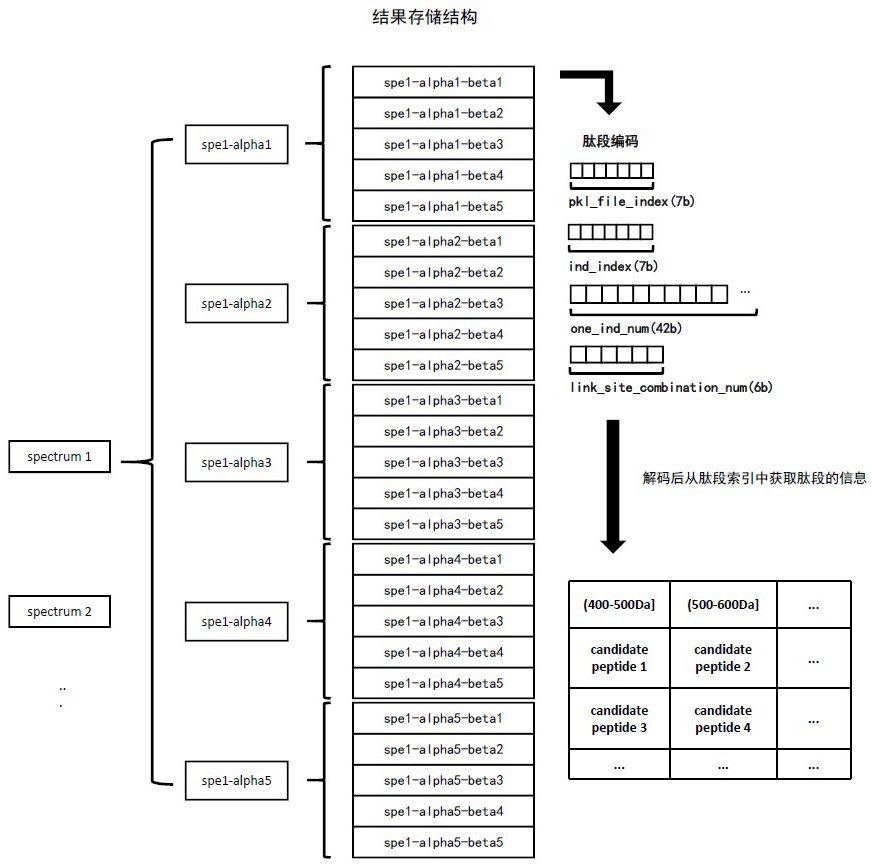
Mass spectrum-based complex cross-linked peptide fragment identification method
An identification method and peptide technology, applied in the field of biological analysis, can solve the problems of no help, wrong identification results, simple cross-linked peptide identification methods cannot be used for complex cross-linked peptide identification, etc., and achieve high accuracy.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0057] Taking the peptide spectrum spectrum of the collected D-DIMER complex crosslinking form using a protein database as an example, the method of the present invention is used to test the mass spectrum data containing D-DIMER, and D-D- DIMER has a spectaccha in complex crosslinking form peptides, such as Figure 5 As shown, LTIGEGQQHLGGAKQAGDV-LTIGEGQQHLGGDV-LTIGEGQQHLGGGDV-LTIGEGQQHLGGAKQAGDV, the underscore is a crosslinking site, and the isopeptide bond is crosslinked to K.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com