Method for detecting ERGIC3 mRNA based on DNA molecules
A technology of DNA molecules and detection probes, which is applied in the field of detection of target ERGIC3mRNA based on DNA molecules, and can solve problems such as inability to detect ERGIC3mRNA
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
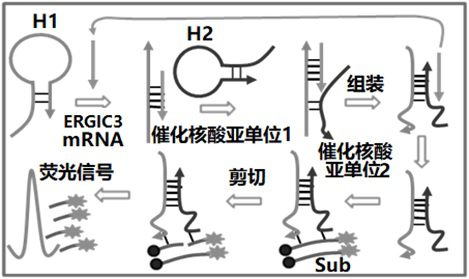
[0048] Such as figure 1 It is a schematic diagram of the method for detecting ERGIC3 mRNA based on DNA molecules of the present invention.
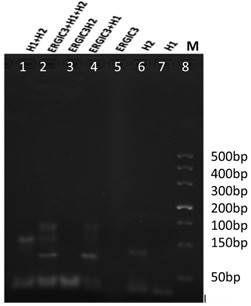
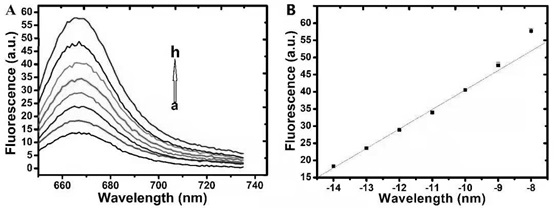
[0049] (1) Feasibility verification results of nucleic acid testing:
[0050] Agarose gel (3%) electrophoresis to verify the ortho-catalyzed hairpin self-assembly reaction, the results are as follows figure 1 shown. Lane 8 is a 500bp DNA marker (Takara), used as a molecular weight reference control for electrophoresis. Lanes 5, 6 and 7 are the corresponding bands of H1, H2 probes and ERGIC3 alone. Lane 4 is the electrophoresis band of the ERGIC3+H1 reaction system, and a lagging band relative to lanes 6 and 7 can be seen, which is due to the assembly of ERGIC3+H1 to form a large molecular weight DNA hybrid, which leads to a slowdown in the electrophoretic movement rate. But from lane 3, it can be seen that ERGIC3 and H2 cannot form DNA hybrids, that is, they do not react. Lane 2 is the electrophoresis band of the assembled chain ERGI...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com