New method for lung adenocarcinoma biomarker screening, prognosis model construction and biological verification
A biomarker, lung adenocarcinoma technology, applied in the field of model construction and biological verification
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
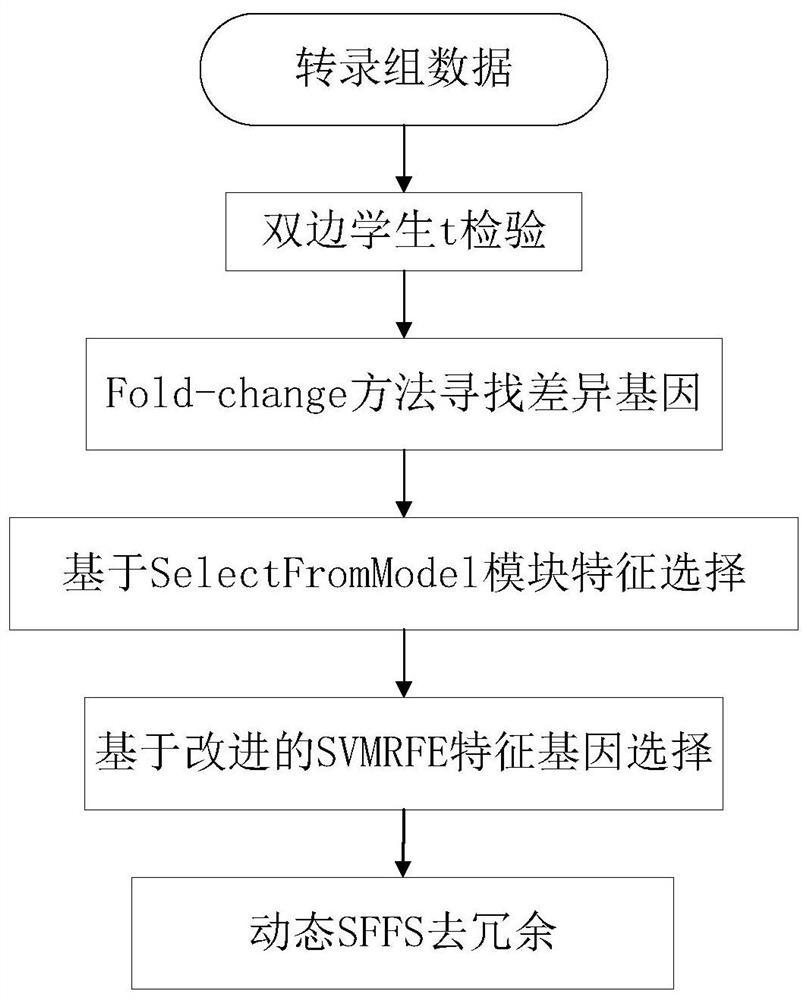
[0050] see Figure 1 to Figure 3 Shown:
[0051] The novel method for lung adenocarcinoma biomarker screening, prognostic model construction and biological verification provided by the present invention comprises the following steps:
[0052] Step 1. Perform data preprocessing on the original gene expression matrix. First, use the matching file to annotate, and change the probe name to the gene name. The process of data preprocessing is as follows:
[0053] 1) Deleting transcriptome signature genes with a deletion rate of more than 20%;
[0054] 2) Delete transcriptome signature genes whose variance is close to 0;
[0055] 3) Use KNN filling method to fill in the transcriptome characteristic genes with a deletion rate of more than 20%;
[0056] 4), use the Z-score method to normalize the expression matrix obtained in 3)
[0057] Finally, a transcriptome expression data matrix containing 57,000 genes and 513 samples was obtained.
[0058] Step 2: Perform biomarker screeni...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com