Automatic analysis method and system for sequencing data of whole genome of bacteria
A whole-genome sequencing and automated analysis technology, applied in sequence analysis, bioinformatics, instruments, etc., can solve problems such as insufficient comprehensiveness, poor user-friendliness, and inability to meet the needs of sequencing data analysis.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0036] The purpose of this example is to provide an automated analysis method for bacterial whole genome sequencing data.
[0037] An automated analysis method for bacterial whole genome sequencing data, comprising:
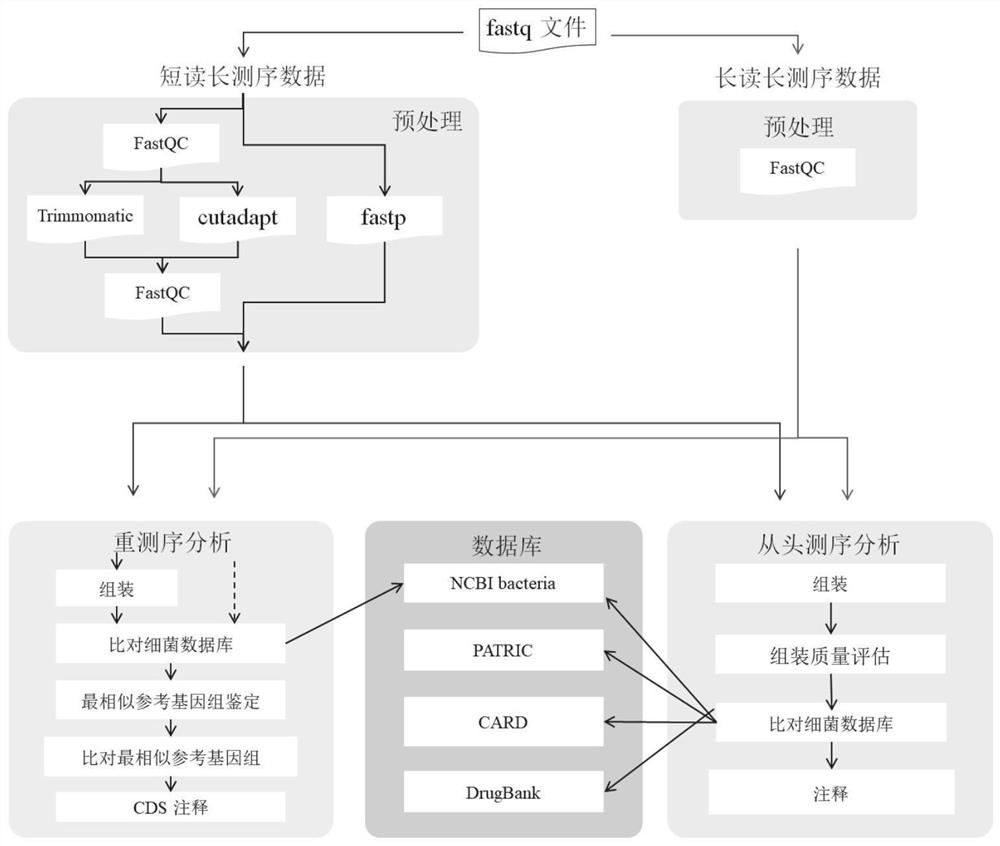
[0038] Obtain bacterial genome sequencing data and determine the type of sequencing data;
[0039] Perform corresponding preprocessing according to the type of sequencing data;
[0040] Perform resequencing analysis and de novo sequencing analysis on the preprocessed sequencing data according to the analysis type selected by the user and the preset tool software and software parameters;
[0041] Enable identification and annotation of bacterial genomes.
[0042] Further, the analysis type selected by the user and the preset tool software and software parameters are saved through a configuration file, and the user realizes related custom settings by modifying the configuration file.
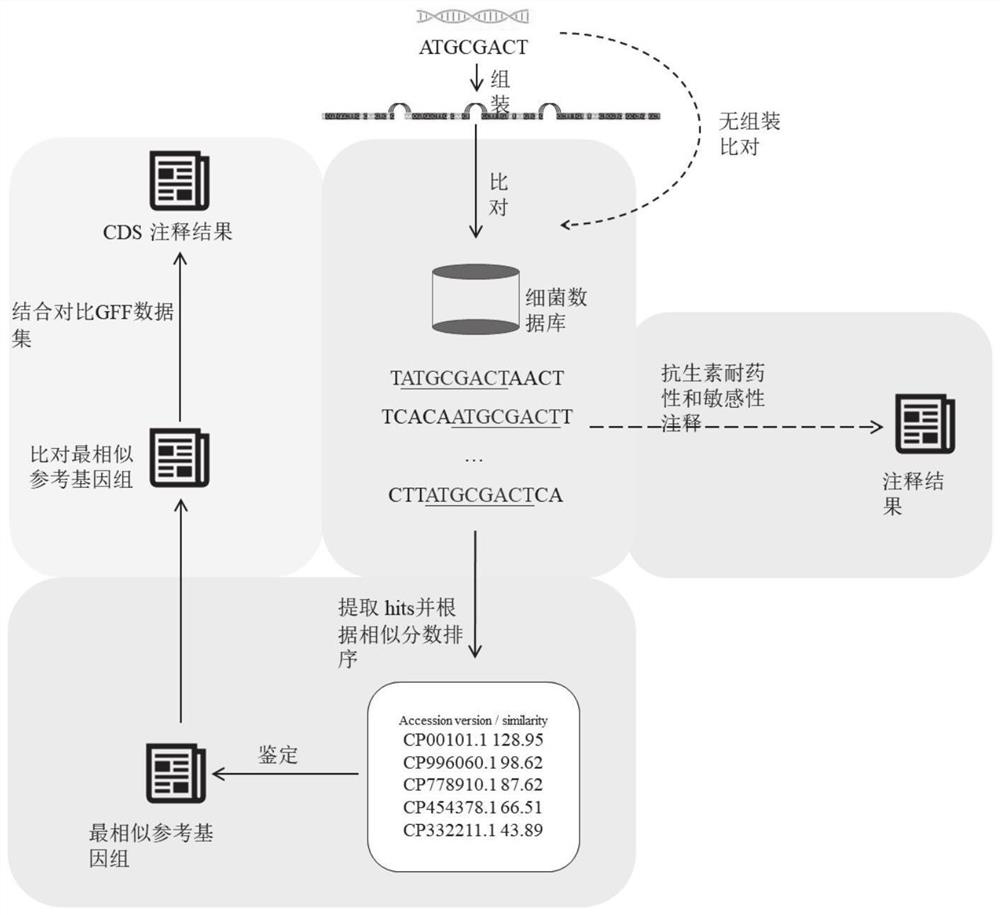
[0043] Further, the resequencing analysis is specifically:
[0044] Assemble...
Embodiment 2
[0070] The purpose of this example is to provide an automated analysis system for bacterial whole genome sequencing data.
[0071] An automated analysis system for bacterial whole genome sequencing data, including:
[0072] A data acquisition unit, which is used to acquire bacterial genome sequencing data and determine the type of sequencing data;
[0073] A preprocessing unit, which is used to perform corresponding preprocessing according to the type of sequencing data;
[0074] The analysis unit is used to perform resequencing analysis and de novo sequencing analysis on the preprocessed sequencing data according to the analysis type selected by the user and the preset tool software and software parameters; to realize the identification and annotation of the whole bacterial genome.
[0075] In further embodiments, there is also provided:
[0076] A computer readable instruction. When the computer instruction is executed by a processor, the method described in the first embo...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com