Construction and application of recombinant saccharomyces cerevisiae for synthesizing carminic acid
A technology of carminic acid and Saccharomyces cerevisiae, which is applied in the field of genetic engineering and bioengineering, and can solve the problems of lack of application
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0049] Example 1: Construction of gene expression cassettes related to carminic acid synthesis pathway
[0050] Using the ZhuI synthetic sequence (nucleotide sequence as shown in SEQ ID NO.3) as a template, design primer pair F1 / R1, carry out PCR amplification with the primer pair, and use Primer Star MasterMix high-fidelity pfu enzyme to carry out, the condition is pre- Denaturation at 95°C, 3min; 30 cycles of amplification, according to 95°C, 10s, 55°C, 10s, 72°C, 30s; extension at 72°C, 5min, the PCR product was purified to obtain the fragment ZhuI; the vector pY26 was used as Template, F2 / R2 was amplified by PCR with primers, and the product was purified. The fragment ZhuI and the vector pY26 were recombined by the Gibson assembly method to obtain a recombinant vector, the recombinant vector was transformed into Escherichia coli JM109, the plasmid was extracted and verified by sequencing, and the correct recombinant vector pY26-zhuI was obtained.
[0051] Using the zhuJ s...
Embodiment 2
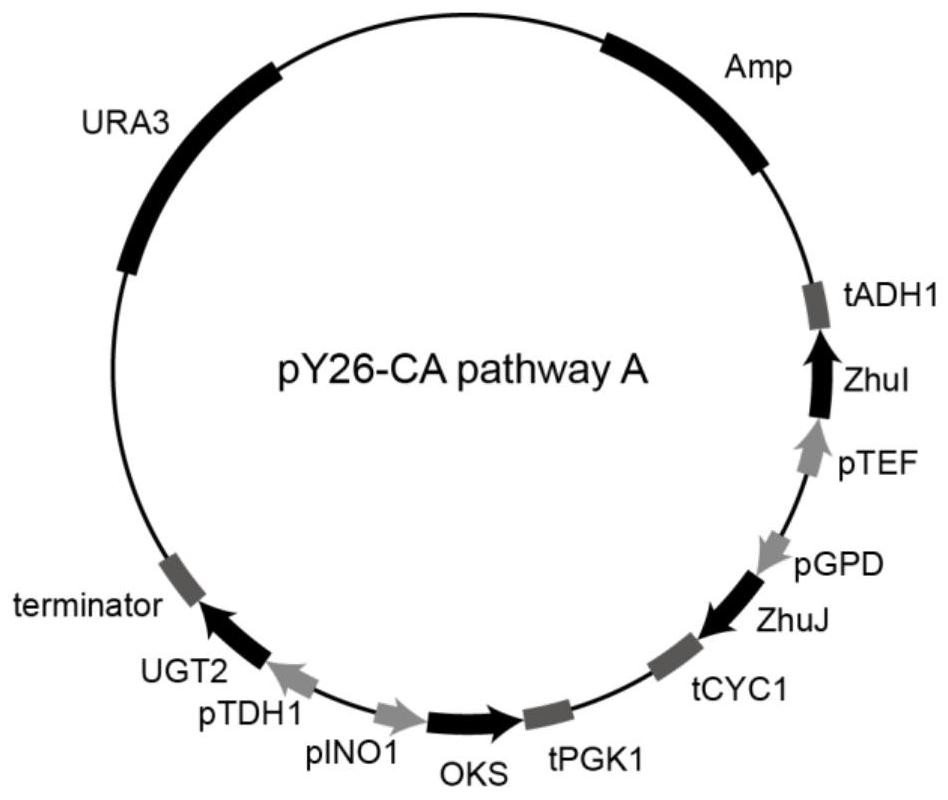
[0059] Example 2: Construction of carminic acid synthesis pathway carrier A
[0060]Using the vector pMD19T-pINO1-OKS-tPGK1 constructed in Example 1 as a template, F19 / R19 was amplified by PCR with primers, and the PCR product was purified to obtain the fragment pINO1-OKS-tPGK1; pMD19T-pTDH1-UGT2-ter-pGAL7 was used as a template, PCR amplification was performed on F20 / R20 with primers, and the PCR product was purified to obtain the fragment pTDH1-UGT2-ter-pGAL7; pY26-zhuI constructed in Example 1 -ZhuJ is used as a template, the primer pair F21 / R21 is used for PCR amplification, and the PCR product is purified to obtain the vector fragment pY26-zhuI-ZhuJ. The fragments pINO1-OKS-tPGK1, pTDH1-UGT2-ter-pGAL7 and the vector pY26-zhuI-ZhuJ were recombined by the Gibson assembly method to obtain the recombinant vector, and the recombinant vector was transformed into Escherichia coli JM109 and sequenced to verify that the correct recombinant vector pY26 was obtained -CA pathway A (...
Embodiment 3
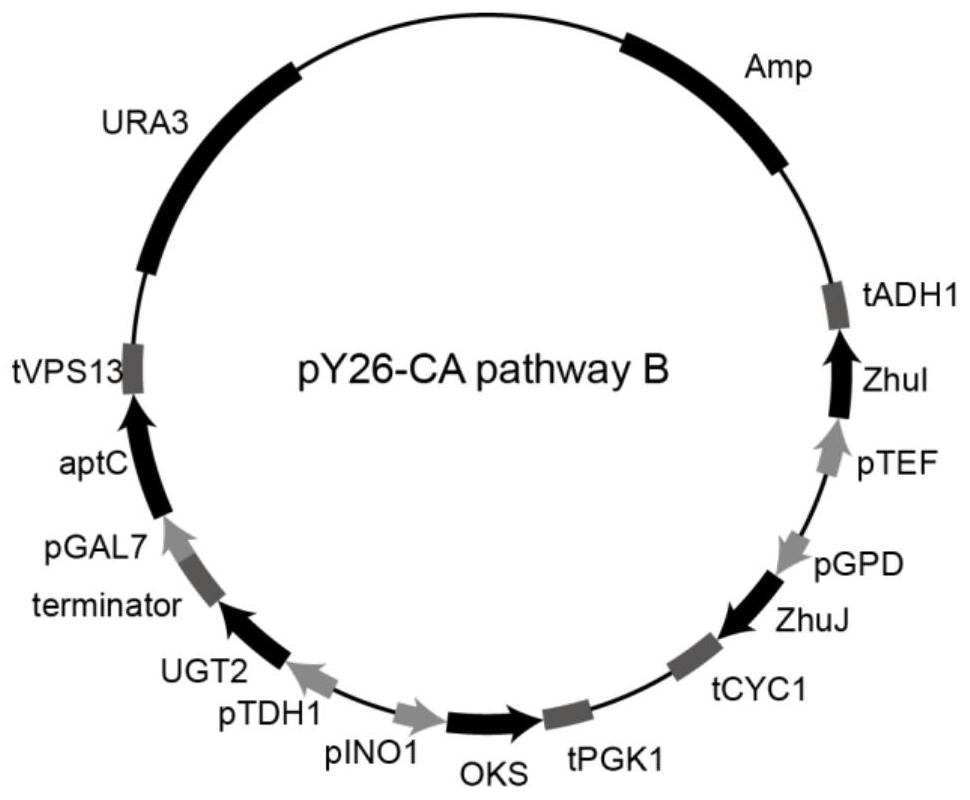
[0063] Example 3: Construction of carminic acid synthesis pathway carrier B
[0064] Using the pMD19T-aptC-tVPS13 constructed in Example 1 as a template, PCR amplification was performed on F22 / R22 with primers to obtain the fragment aptC-tVPS13; using pY26-CA pathway A as a template, PCR amplification was performed on F23 / R23 with primers The linearized vector pY26-CA pathway A was obtained. The fragment aptC-tVPS13 and the vector pY26-CA pathway A were recombined by the Gibson assembly method to obtain the recombinant vector, and the recombinant vector was transformed into Escherichia coli JM109 and sequenced to verify that the correct recombinant vector pY26-CA pathway B (pY26-zhuI -ZhuJ-pINO1-OKS-tPGK1-pTDH1-UGT2-ter-pGAL7-aptC-tVPS13) (see figure 2 ).
[0065] Primers used in Table 3
[0066] Primer Sequence 5'-3' (the underlined part is the homology arm region) F22 TCCCTCAAAA ATGACTTTGCCAGTTTTGATTATTGGTG
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com