Chromatin imaging method and chromatin imaging system based on Type I-F CRISPR/Cas
An imaging system, chromatin technology, applied in the field of molecular biology
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0100] The sequence optimization of embodiment 1 PaeCascade
[0101] The PaeCascade sequence is derived from the type I-F CRISPR complex of Pseudomonas aeruginosa. Jennifer A. Doudna's team published an article entitled "RNA-guided complex from a bacterial immune system enhances target recognition throughseed sequence interactions" in "Proc Natl Acad Sci" in 2011, which reported that PaeCascade can bind dsDNA sequences in vitro. Its source strain is Pseudomonas aeruginosa.
[0102] Since the sequences of the four proteins (Cas8f1, Cas5f1, Cas7f1, and Cas6f) at the PaeCascade site are regulated by prokaryotic polycistrons, they are prokaryotic preferred codon coding sequences (SEQ ID NO.1). Therefore, in order to promote the expression of PaeCascade in mammalian cells, its sequence characteristics should be optimized. There are three principles for the transformation: 1) Split out individual proteins, each of which is optimized for mammalian codons; 2) Remove unexpected eukar...
Embodiment 2
[0105] Embodiment 2 constructs recombinant plasmid
[0106] 1. Construct the recombinant plasmid pxCMV-hCas5f1-PGK-hCas8f1 (sequence shown in SEQ ID NO.7)
[0107] The AscI and MluI restriction sites and the NotI and HindIII restriction sites were connected to the improved pxCMV plasmid respectively, hCas8f1 and hCas5f1 containing the NLS sequence (PKKKRKV) were connected and the pxCMV-hCas5f1-PGK-hCas8f1 plasmid (SEQ ID NO .7).
[0108] The improved pxCMV used was derived from the px601 plasmid, in which an AscI restriction site was added behind the CMV promoter, and the PGK promoter and NotI restriction site were constructed by inserting the original KpnI and HindIII sites.
[0109] 2. Construct the recombinant plasmid pxCMV-hCas6f-PGK-hCas7f1 (sequence shown in SEQ ID NO.8):
[0110] Linked to the improved pxCMV plasmid through AscI and MluI restriction sites and NotI and HindIII restriction sites respectively, hCas7f1 and hCas6f containing NLS sequence (PKKKRKV) were con...
Embodiment 3
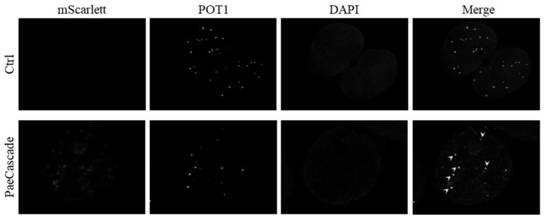
[0121] Example 3 Construction of PaeCascade-mScarlett Chromatin Imaging System
[0122] The PaeCascade-KRAB transcriptional repression system includes:
[0123] (1) pxCMV-hCas5f1-PGK-hCas8f1 plasmid (SEQ ID NO.7),
[0124] (2) pxCMV-hCas7f1-mScarlett-PGK-hCas6f plasmid (SEQ ID NO.9),
[0125](3) The crRNA expression plasmid is pLenti-DR(hCas6f)-EV (SEQ ID NO.10).
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com