Method for screening small molecule peptidomimetic inhibitor and application of small molecule peptidomimetic inhibitor
An inhibitor, small molecule technology, applied in the field of medicine, can solve the problem of lack of specific antiviral drugs
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0038] Embodiment 1 Processing of SARS-Cov-2-RdRp protein crystal structure
[0039] The three-dimensional crystal structure of RdRp protein can be searched and downloaded from the PDB data, the PDB code is 7BV2, and the resolution of the crystal structure is It is the nsp12-nsp7-nsp8 complex, including the RNA chain and the phosphate form of remdesivir structure. Use the "pretein preparation wizard" of Maestro software to correct the chemical bond sequence of the 7BV2 structure, add hydrogen, deal with metal ions, fill in missing atoms and amino acid residues, delete redundant molecules, etc., and then perform protein preparation under OPLS force field conditions Energy optimization, and finally use it as an acceptor for molecular docking.
Embodiment 2
[0040] Example 2 drug binding site
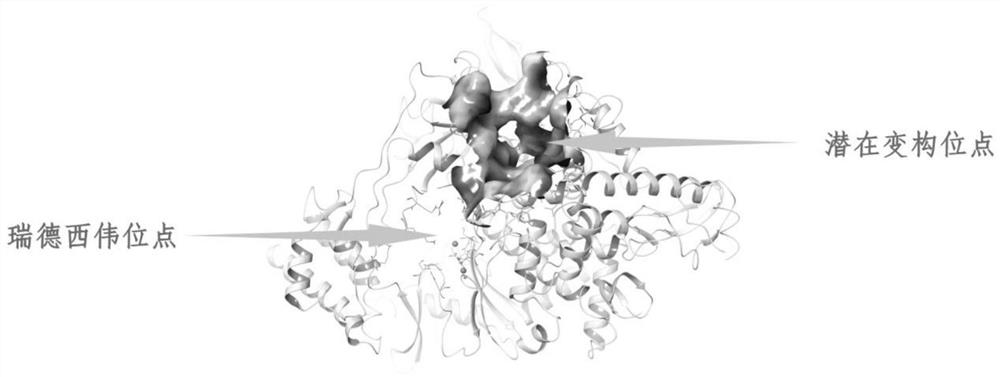
[0041] 7BV2 is a complex complex composed of nsp12-nsp7-nsp8 complex and template primer RNA, and only nsp12 peptide chain is retained when processing protein. In order to detect potential binding sites of nsp12, this example uses the FTmap (http: / / ftmap.bu.edu / login.php) online platform and the site map section in Maestro. The results show that there are two main binding sites, one is the binding site of remdesivir metabolic active substances, and the key amino acid residues that make up the pocket are LYS545, SER682, ASP623, THR687, SER759, ASP760 and ASN691, etc.; The binding site (potential allosteric site) next to the site, the key amino acid residues that make up the pocket are PHE165, PRO169, ASP170, LEU172, ARG173, THR246, LEU247, ARG249, TYR265, THR319, PRO323, THR324, ARG349 , LEU389, ASP390, LYS391, THR393, THR394, PHE396, ARG457, TYR458, LEU460, PRO461, THR462 and PRO677, etc. The positions of the two binding sites are as im...
Embodiment 3
[0042] The selection of embodiment 3 compound library
[0043] Recent studies have shown that the anti-SARS-CoV-2 virus is basically a peptidomimetic compound. In this example, a compound library based on bacterial and fungal metabolites was established with the help of the compound library provided by npatla, which contains a large number of peptide-like compounds. Molecular docking analysis of RNA-dependent RNA polymerase (RdRp) inhibitors was carried out by means of computer-aided drug design (CADD) method.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com