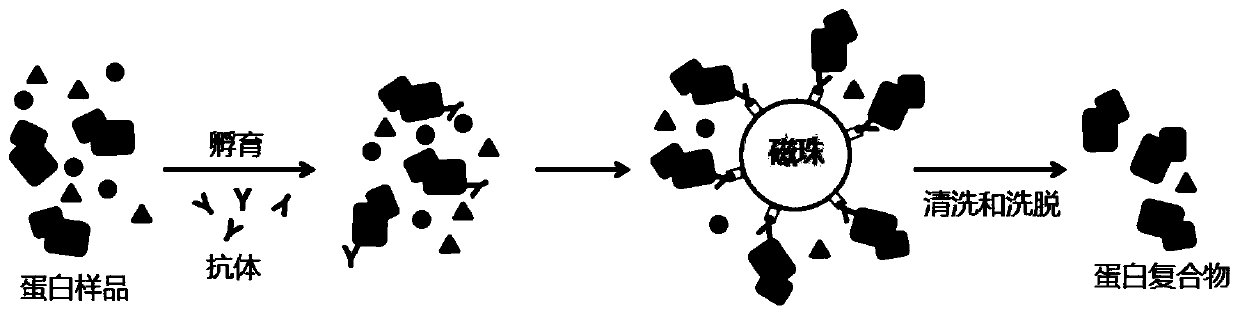
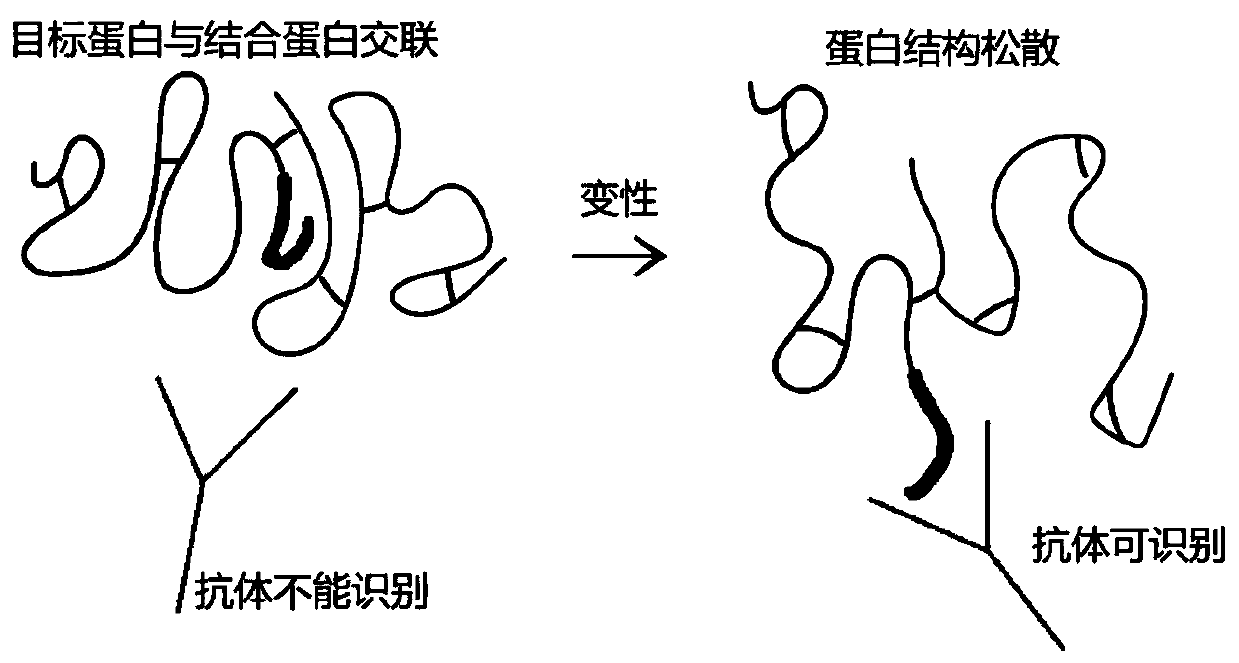
Method of detecting protein complex in biological sample
A detection method and complex technology, applied in the fields of biochemical analysis and protein analysis, can solve problems such as lack of antibodies, and achieve the effect of enriching sources and reducing steric hindrance effects.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0052] In this embodiment, Hela cells are taken as an example, heat shock protein 90 is used as the target protein, and the heat shock protein complex is detected in situ in the cell, and the specific operation is as follows.
[0053] (1) Covalent crosslinking
[0054] Will 1×10 7 Resuspend Hela cells in 450 μl phosphate buffer, add 50 μl 50 mM DSSO stock solution to make the final concentration 5 mM, place at 4 ° C for 1 h, add 1 M NH 4 HCO 3 The final concentration was 20mM to terminate the reaction, and centrifuged at 500×g for 5min to obtain cross-linked cells.
[0055] (2) Denaturation treatment
[0056] Divide the cross-linked cells into two parts, directly add 8M urea lysate 3 times the volume of the cells to one part of the cross-linked cells, sonicate on ice for 5 minutes (on 2s and stop for 3s), and extract denatured protein to obtain denatured protein solution (hereinafter Referred to as the treatment group according to the method of the present invention), anot...
Embodiment 2
[0066] In this embodiment, A549 cells are taken as an example, glyceraldehyde-3-phosphate dehydrogenase (GAPDH) is used as the target protein, and the GAPDH protein complex is detected in the cell lysate, and the specific operation is as follows.
[0067] (1) Covalent crosslinking
[0068] Will 1×10 7 A549 cells were resuspended in 450 μl phosphate buffer, added non-denaturing cell lysate (25mM HEPES, 150mM NaCl, 1% Triton X-100, pH 7.4) to extract the whole protein solution of the cells, and quantitatively obtained 4.4mg / ml protein solution 0.5 ml. Then add 50mM DSBU stock solution to make the final concentration 5mM, place it at 4°C for 1h, add 1M NH 4 HCO 3 The final concentration was 20mM to stop the reaction, and centrifuged at 500×g for 5min to obtain the cross-linked protein solution.
[0069] (2) Denaturation treatment
[0070] The cross-linked protein solution was divided into two parts and diluted to 2.2 mg / ml with non-denaturing cell lysate (25 mM HEPES, 150 mM...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com