In-situ hybridization probe primer, probe and mapping method of muscle satellite cell of sebastes schlegeli
A muscle satellite cell and in situ hybridization technology is applied in the field of in situ hybridization probe primers for muscle satellite cells of Xu's flat scorpionfish, which can solve the problems of unclear mechanism and the like
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
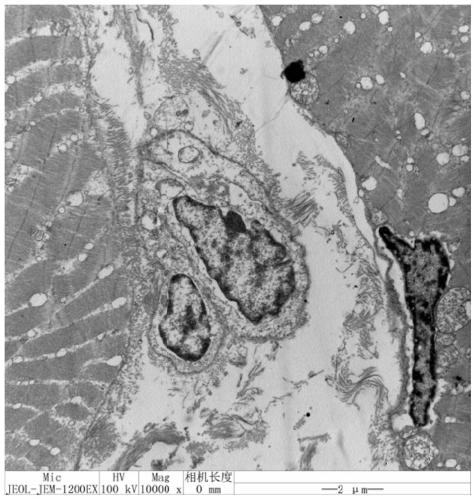
[0024] A method utilizing transmission electron microscopy to locate muscle satellite cells in the muscle of the scorpionfish Xu's, the specific steps of the method are as follows:
[0025] 1. Sampling and washing. Take a 1-year-old flat scorpionfish with a clean and sterile blade above the lateral line, close to the skeletal muscle in the middle of the dorsal fin, cut into small pieces about 1mm wide and 2-3mm long, and rinse with excess RNase-free 1xPBS buffer. The washed muscle samples were stored overnight at 4°C in 2.5% glutaraldehyde;
[0026] 2. After 0.1mol L -1 After rinsing with phosphate buffer, transfer to 1% osmic acid and fix at 4°C for 3h.
[0027] 3. Dehydration. 50% ethanol, 15-20min; 70% ethanol, 15-20min; 90% ethanol, 15-20min; 90% ethanol, 90% acetone (1:1) 15-20min; 90% acetone, 15-20min. All the above operations were carried out at 4°C. Dehydrate in 100% acetone at room temperature three times, each time for 15-20min.
[0028] 4. Embedding. Pure ac...
Embodiment 2
[0031] Embodiment 2 A probe primer for in situ hybridization of Xu's flat scorpion muscle satellite cells, the primer sequence is as follows: F: 5'-GGACAGGCCTACTAAAG-3', R: 5'-CCATGACAAACAGGAAC-3'; synthesized using the above primers Probe, the probe sequence is:
[0032]GGACAGGCCTACTAAAGCCCCCCCACCCCACCCCTAGGTCGCACCCTGCGCCACTCTCCTCCGCCCTCTGCCACCCTCCTCCATGCTCCATCAAGTCTGGACTCCCAACTCCAGCTCGCTCTGCAGCCGGGCCATGACATCAGCGCACAGACACGCAGCCTCGCTGAGAGAAAGTGACGCTCGCGAGTGTGACAAAAGCACTTCGGGGGAATGGAACTAAAGTTTTCACGCCATTGTCAACCACCAGGCGGCTCTGCACCCTCTCTTCTCAACCTTTTAGGCGTCTCTGCCTGTTCCCTGTACTCTCCTTCCTCCAGCCCCAAACCCTGCTTTGTTTTCCCCCAGAAGGCCAGACGGGATCCAAGTTCAACCGAGGATGGTCGGCCGCCTTGGCCCGGGTTCCTGTTTGTCGTGG
[0033] Utilize above-mentioned probe to carry out in situ hybridization localization method to Xu's flat scorpion muscle satellite cell, concrete steps are as follows:
[0034] 1) In situ hybridization sample fixation. Take the skeletal muscle sample from the same position on the other side of the s...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com