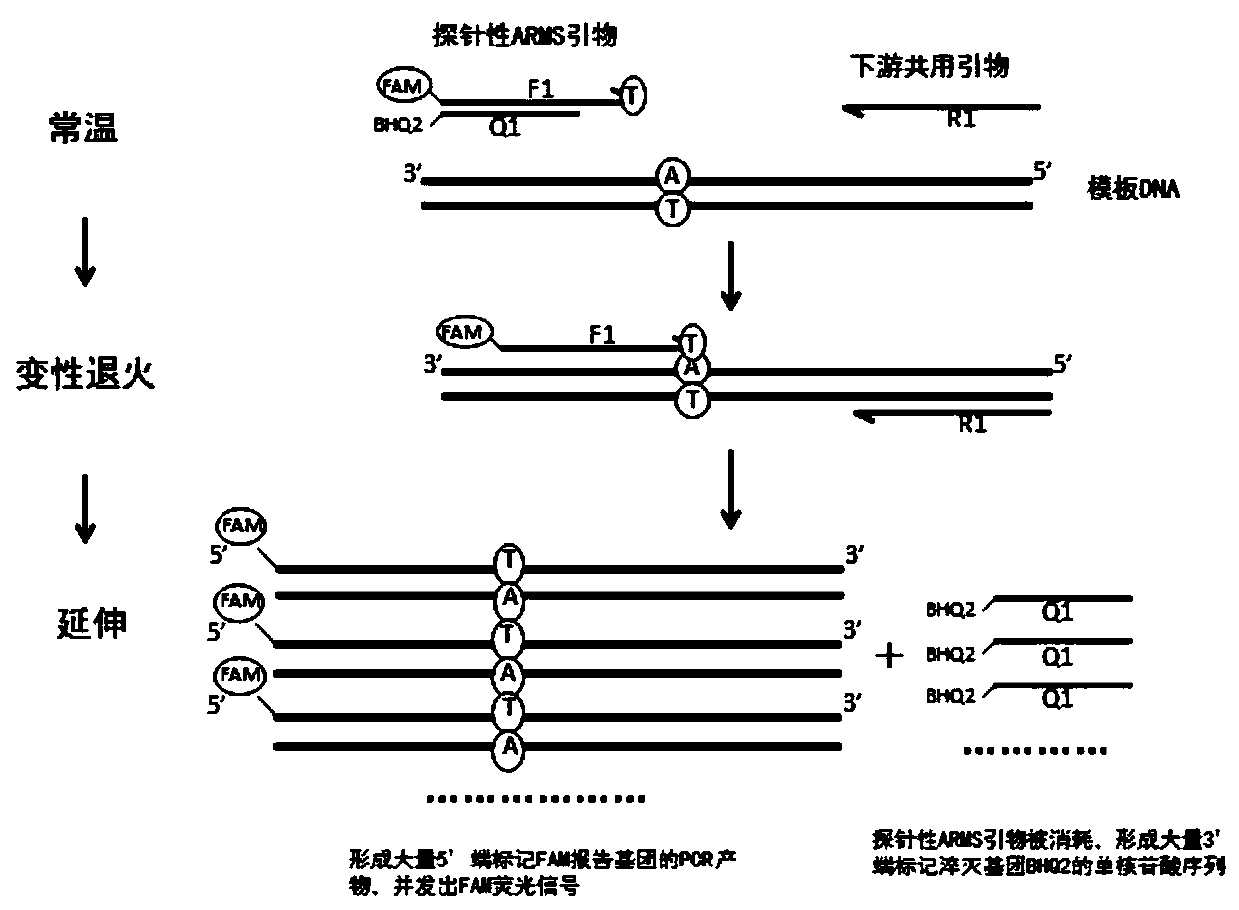
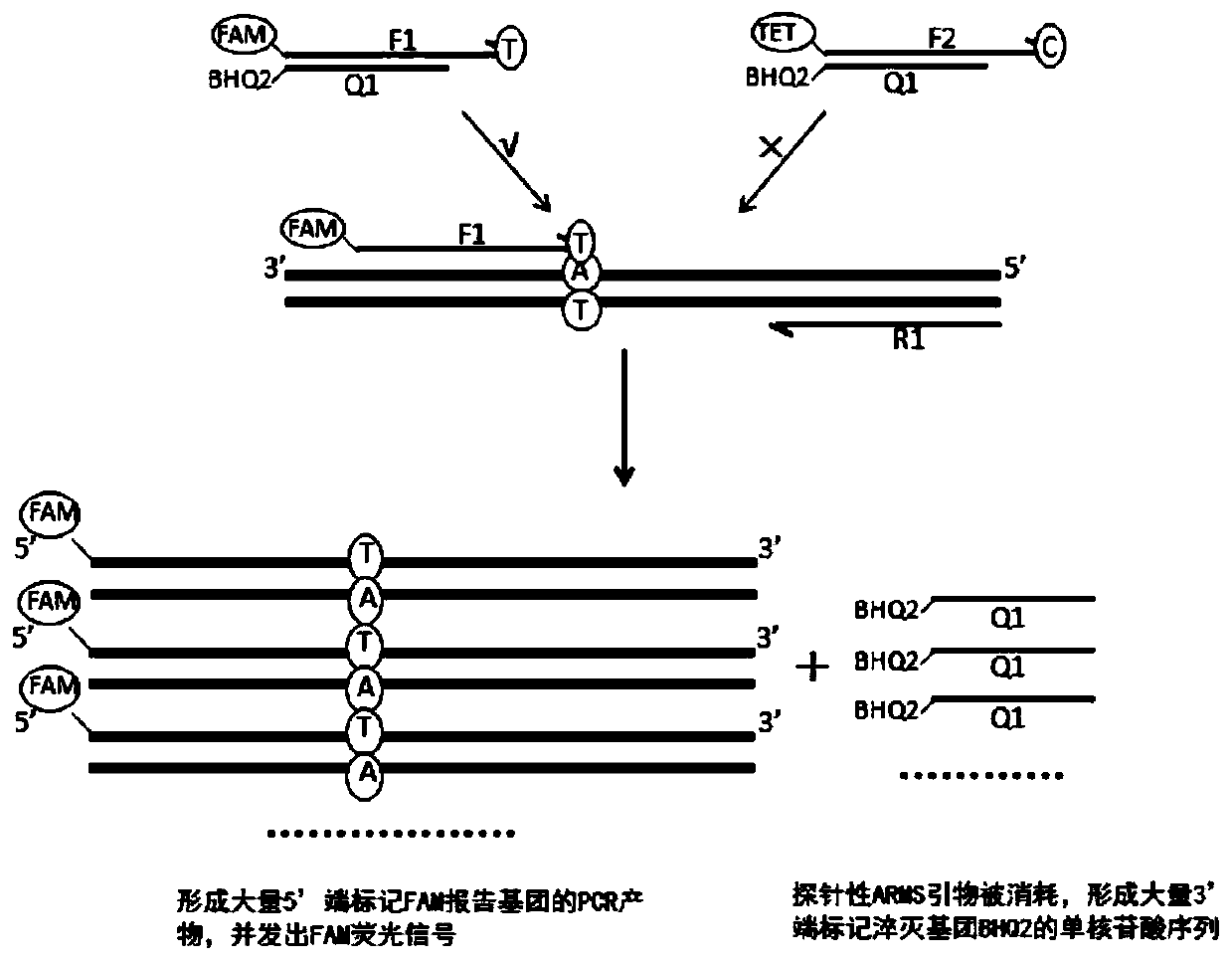
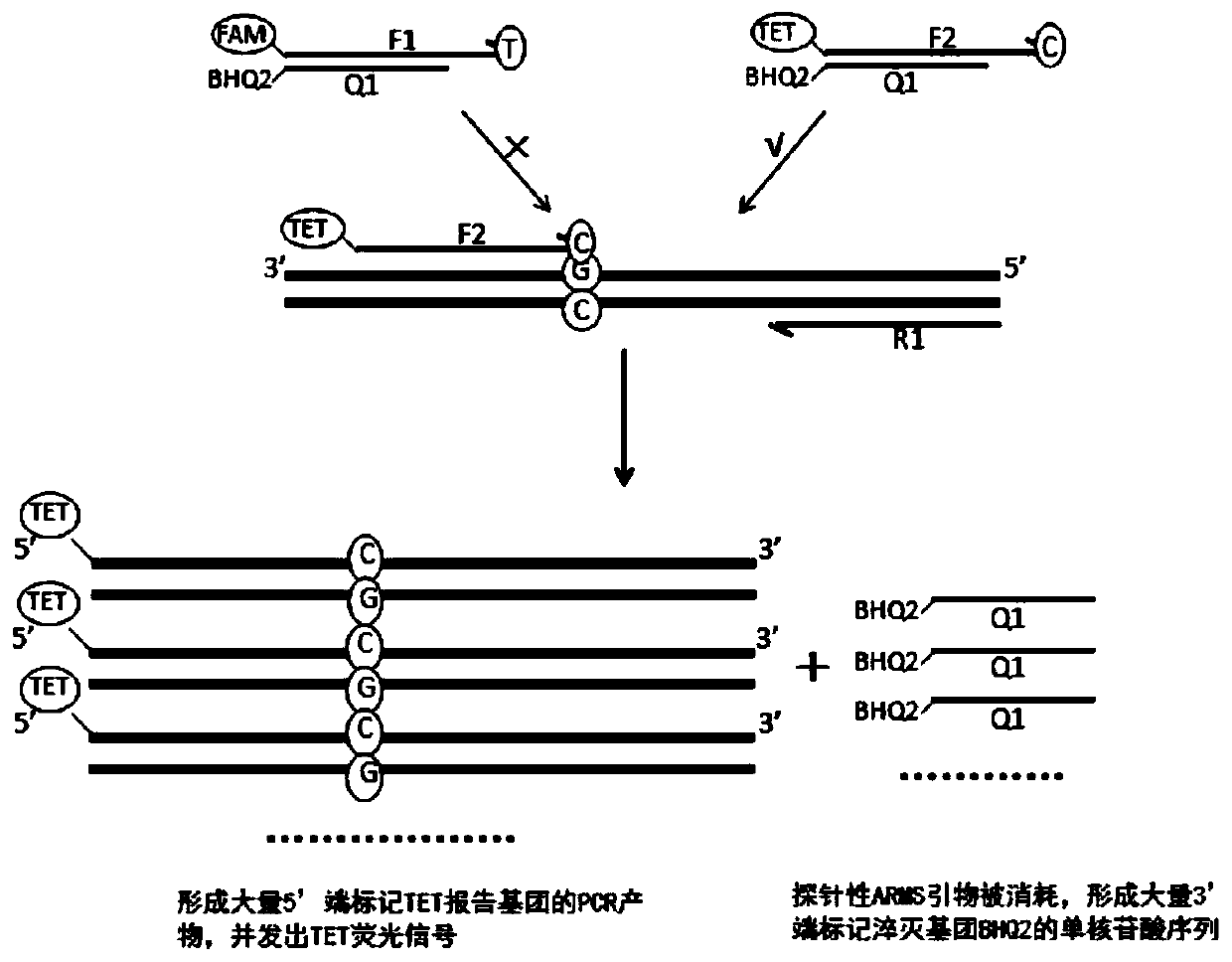
Kit, method and application for detecting gene polymorphism based on shared primer probe
A gene polymorphism and kit technology, applied in the field of PCR detection of gene polymorphism, can solve the problems of detection time up to 4 to 5 hours, cumbersome operation, special equipment, PCR product contamination, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0135] Example 1 Design optimization of primers, optimization screening of markers
[0136] 1. The design and screening of wild-type and mutant-type probe ARMS primers, the specific steps are as follows:
[0137] 1) Preliminarily design a series of ARMS primers for CYP2C19*2, CYP2C19*3, CYP2C19*17 wild-type and mutant loci, with a Tm value of 58-62°C, and the sequences are shown in Table 1:
[0138] Table 1. Preliminary design of CYP2C19*2, CYP2C19*3, CYP2C19*17 wild-type and mutant ARMS primers
[0139]
[0140]
[0141]
[0142] 2) CYP2C19*2 wild-type ARMS primers, downstream primers, and SYBR-premix buffer to prepare PCR reaction solution, respectively detect 5000 copy / μl of CYP2C19*2 wild-type plasmid and mutant plasmid; CYP2C19*2 mutant ARMS primers, downstream primers , SYBR-premix buffer to prepare PCR reaction solution, respectively detect 5000 copy / μl of CYP2C19*2 wild-type plasmid and mutant plasmid; CYP2C19*3 wild-type ARMS primer, downstream primer, SYBR-...
Embodiment 2
[0180] Example 2 Primer preparation and kit assembly
[0181] 1. Synthesis and fluorescent labeling of probe-based ARMS primers, quenching probes, and downstream primers in this application
[0182] According to Example 1, optimize and screen the probe ARMS primers, quencher probes, downstream primers and markers, and perform primer synthesis and fluorescent labeling. The specific markers are CYP2C19*2 wild-type probe ARMS primers 5′-end labeled FAM, CYP2C19*2 mutant probe ARMS primer 5′-end labeled TET, CYP2C19*2 quencher probe 3′-end labeled BHQ2, CYP2C19*2 downstream primer; CYP2C19*3 wild-type probe ARMS primer 5′-end labeled ROX , CYP2C19*3 mutant probe ARMS primer 5′-end labeled CY5, CYP2C19*3 quencher probe 3′-end labeled BHQ2, CYP2C19*3 downstream primer; CYP2C19*17 wild-type probe ARMS primer 5′-end labeled FAM, CYP2C19*17 mutant type probe ARMS primer 5′ end labeled TET, CYP2C19*17 quenching probe 3′ end labeled BHQ2, CYP2C19*17 downstream primer.
[0183] The spec...
Embodiment 3
[0224] Embodiment 3 detection sensitivity
[0225] Use the kit a for detecting human CYP2C19 gene polymorphisms based on shared primer probes prepared in Example 2 and the kit for detecting CYP2C19 gene polymorphisms based on ARMS primers + TaqMan probes b, and the detection of CYP2C19 gene polymorphisms based on MGB probes morphological kit c comparative test, the specific comparative test steps are as follows:
[0226] Step 1. Prepare sensitivity templates, including CYP2C19*2 homozygous wild type 2ng / μl, 1ng / μl, 0.5ng / μl, CYP2C19*2 homozygous mutant 2ng / μl, 1ng / μl, 0.5ng / μl, CYP2C19* 2 heterozygous 2ng / μl, 1ng / μl, 0.5ng / μl; CYP2C19*3 homozygous wild type 2ng / μl, 1ng / μl, 0.5ng / μl, CYP2C19*3 homozygous mutant 2ng / μl, 1ng / μl μl, 0.5ng / μl, CYP2C19*3 heterozygous 2ng / μl, 1ng / μl, 0.5ng / μl; CYP2C19*17 homozygous wild type 2ng / μl, 1ng / μl, 0.5ng / μl, CYP2C19*17 pure Synzygous mutant 2ng / μl, 1ng / μl, 0.5ng / μl, CYP2C19*17 heterozygous 2ng / μl, 1ng / μl, 0.5ng / μl;
[0227] Step 2, prepar...
PUM
| Property | Measurement | Unit |
|---|---|---|
| melting point | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com