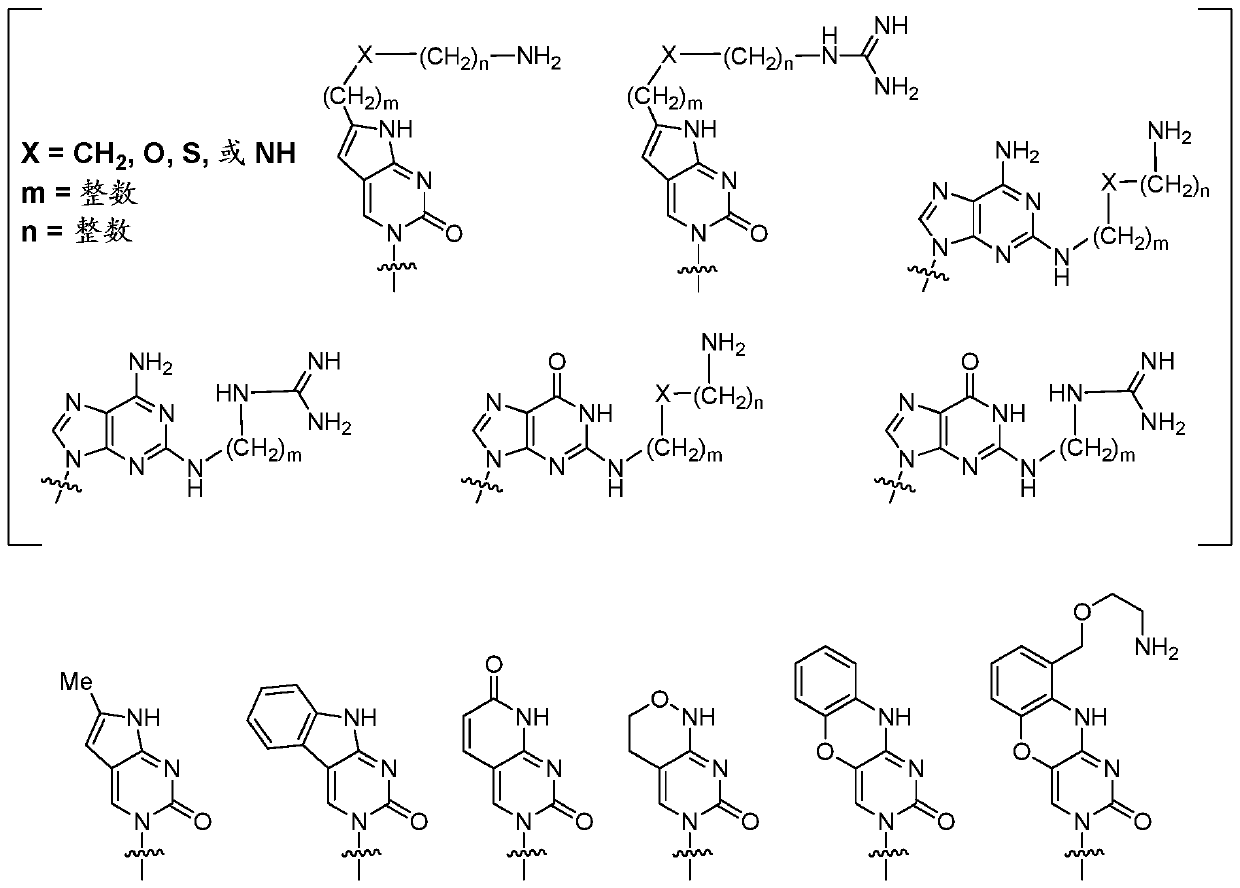
Scn9a antisense pain killer
一种药学、药用盐的技术,应用在非中枢性止痛剂、重组DNA技术、DNA / RNA片段等方向
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0343] Example 1. Exon skipping induced by "ASO 7" in PC3 cells (A)
[0344] "ASO 7" designated in Table 1 is a 14-mer ASO that binds complementary to a 14-mer sequence at the 3' splice site spanning the junction of intron 3 and exon 4 within the human SCN9A pre-mRNA . 14-mer target sequence within the 3' splice site at [(5' → 3') uugu guuuag ┃ GUACACUU UU] are marked as "bold" and "underlined". "ASO 7" has a 6-mer complementary overlap with intron 3 and an 8-mer complementary overlap with exon 4.
[0345] Given that PC3 cells (Cat. No. CRL1435, ATCC) are known to express abundant human SCN9A mRNA [ Br. J. Pharmacol vol 156, 420-431 (2009)], "ASO 7" was evaluated for its ability to induce exon 4 skipping in PC3 cells as follows.
[0346] [Cell culture and ASO treatment] PC3 cells grown in a 60 mm dish containing 5 mL of F-12K medium were treated with "ASO 7" at 0 (negative control), 1, 10 or 100 zM.
[0347] [RNA extraction and cDNA synthesis by one-step PCR] After ...
Embodiment 2
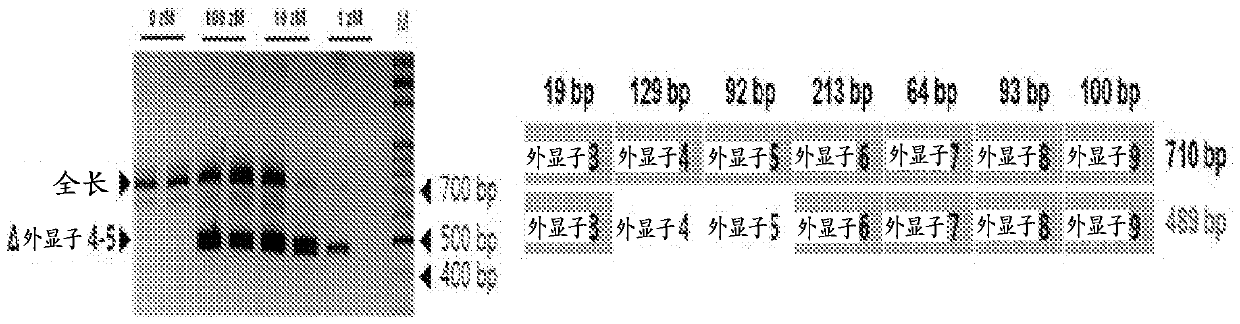
[0351] Example 2. Exon skipping induced by "ASO 7" in PC3 cells (B)
[0352] The ability of "ASO 7" to induce "exon 4" skipping in PC3 cells was evaluated as in "Example 1", unless otherwise indicated. In this experiment, PC3 cells were treated with "ASO 7" at 0 (negative control), 1, 10, 100 and 1,000 aM for 24 hours. Nested PCR reactions were performed against a set of primers [exon 3 / 6_forward: (5' → 3') GGACCAAAAATGTCGAGCCT; and exon 9_reverse: (5' → 3') GCTAAGAAGGCCCAGCTGAA], the The primers described above were designed to amplify products with the sequence at the junction of exon 3 and exon 6.
[0353] The primer sequence for "exon 3 / 6_forward" more selectively recognizes the junction of "exon 3 and exon 6" than the "exon 3 and exon 6" found in the full-length SCN9A mRNA The junction of child 4". Primer sequences were designed to detect SCN9A splice variants missing exons 4–5 more sensitively than full-length SCN9A mRNA. Such exon-skipping primers will help detect m...
Embodiment 3
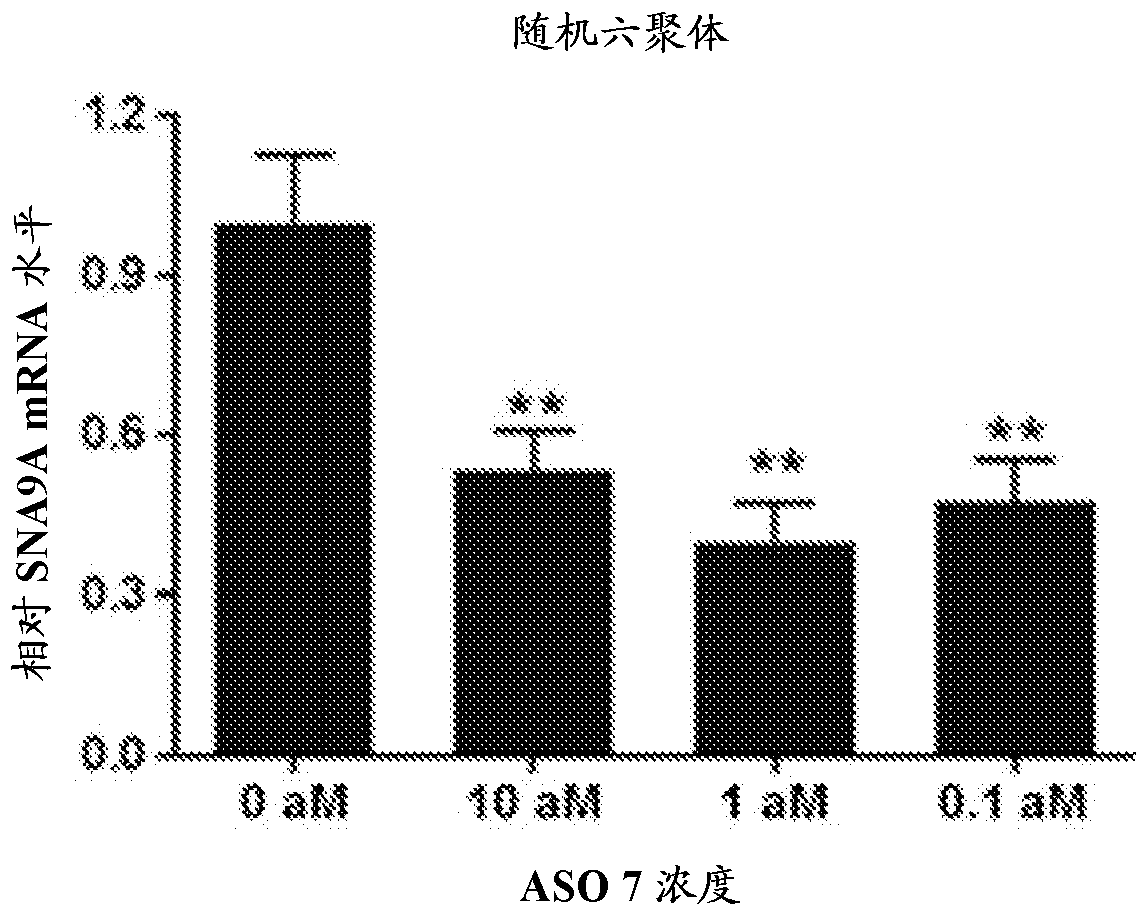
[0355] Example 3. qPCR of SCN9A mRNA in PC3 cells treated with "ASO 7" (A)
[0356] "ASO 7" was evaluated by SCN9A nested qPCR for its ability to induce changes in human SCN9A mRNA levels in PC3 cells as described below.
[0357] [ASO Treatment] PC3 cells were treated with "ASO 7" at 0 (negative control), 0.1, 1 or 10 aM (2 culture dishes for each ASO concentration).
[0358] [RNA extraction and cDNA synthesis by one-step PCR] After incubation with "ASO 7" for 24 hours, total RNA was extracted and subjected to one-step PCR amplification as described in "Example 1".
[0359] [Nested qPCR amplification] Dilute the cDNA solution 100-fold, and make 1 μL of each diluted PCR products were subjected to a set of exon-specific primers for [exon 3_forward: (5'→3') TGACCATGAATAACCCAC; and exon 4_reverse: (5'→3')GCAAGGATTTTTACAAGT] 20 μL real-time PCR reaction. qPCR reactions were performed with a TaqMan probe of [(5' → 3') 5,6-FAM-GGACCAAAA-Zen-ATGTCGAGTACAC-3IABkFQ], which targets ex...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com