Method for efficiently purifying and identifying cyclic RNA based on second-generation Illumina sequencing platform
A sequencing platform and circular technology, applied in the field of molecular biology to improve removal efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0036] Example 1 Extraction of total RNA
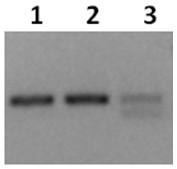
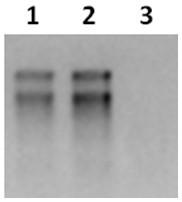
[0037] Take 100-150 mg of the ground sample, use the RNAprep Pure Polysaccharide Polyphenol Plant Total RNA Extraction Kit (Tiangen, Cat.#DP441) to extract the total RNA of the sample, and then use the DNA digestion enzyme gDNA Eraser to remove the genomic DNA. box instructions. Detect RNA bands by 1% agarose gel electrophoresis. If the brightness of 28S rRNA is 1.5-2 times that of 18S rRNA, it means that the RNA is not degraded. RNA concentration and absorbance were measured using a NanoDrop 2000c UV-Vis spectrophotometer, and the A260 / 280 was between 1.8-2.0, indicating that the purity of the RNA sample was high. The Agilent 2100 bioanalyzer was used to further detect the integrity number (RNA integrity number, RIN) of the RNA sample. If the RIN value was greater than 7.5, the sequencing requirements were met. Take 10ug of good quality total RNA for library construction experiments.
Embodiment 2
[0038] Example 2 Removal of polyadenylated mRNA in total RNA
[0039] (1) Oligo-dT Dyna magnetic beads pretreatment
[0040] 1) Use Dynabeads mRNA Purification kit (Invitrogen, Cat.61006) to thoroughly resuspend Oligo-dT Dyna magnetic beads, evenly pipette 200 μL into a new RNase-free tube, and place the tube on the magnetic stand for 1 -2min, the magnetic beads will gather at the magnetic end, transfer the supernatant with a pipette, discard it, and keep the tube on the magnetic stand at the same time.
[0041] 2) Resuspend Oligo-dT Dyna magnetic beads with 200 μL Binding Buffer, remove the tube from the magnetic stand, gently pipette several times to mix, put it back on the magnetic stand, let it stand for 1-2min, carefully transfer the supernatant, and discard . Repeat this step twice.
[0042] 3) Resuspend the Oligo-dT Dyna magnetic beads in 200 μL Binding Buffer again, and divide them equally into RNase-free tube 1 and tube 2, 100 μL in each tube.
[0043] (2) Removal...
Embodiment 3
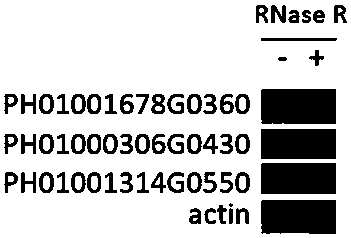
[0059] Example 3 Ribosomal RNA removal
[0060] (1) Ribosomal RNA and probe hybridization
[0061] 1) Set the temperature of two water baths to 70°C and 37°C respectively
[0062] 2) Add to 1.5mL RNase-free tube
[0063]
[0064] The ribosome probe and Hybridization buffer are provided in the RiboMinus™ Plant Kit for RNA-Seq (Invitrogen, A1083808).
[0065] 3) Water bath at 70°C for 5 minutes to denature the RNA.
[0066] 4) Transfer the sample to a 37°C water bath for 30 minutes to slowly cool the RNA down to 37°C
[0067] 5) During the cooling process, pretreat the magnetic beads.
[0068] (2) Magnetic bead pretreatment
[0069] 1) Vortex the magnetic beads in the vial, pipette 750μL into a 1.5mL tube, place on the magnetic stand for 1min, remove the supernatant
[0070] 2) Add 750 μL DEPC water to wash, slowly vortex to resuspend, place on the magnetic stand for 1min, remove the supernatant, repeat this step
[0071]3) Add 750μL Hybridization buffer and slowly vor...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com