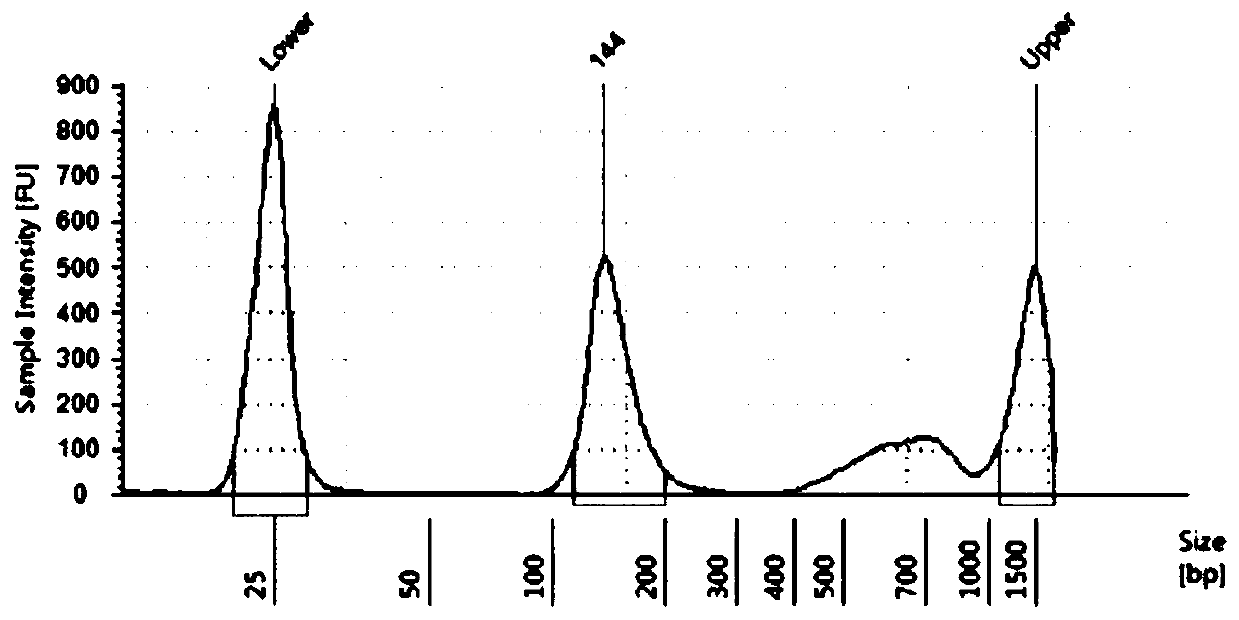
Detection method and kit for mutation rates of lung cancer mutation sites
A detection method and mutation site technology, which is applied in the fields of medicine and biology, can solve the problems of low detection sensitivity, increase in the total amount of cfDNA extraction, and high cost, and achieve the effect of high sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0066] Example 1 cfDNA extraction optimization
[0067] The content of ctDNA in peripheral blood is very low, the ratio is about 1 / 1000, so it is very important to ensure the quality of plasma cfDNA extraction.
[0068] Preferably, the detailed steps of the cfDNA extraction scheme are as follows:
[0069] 1. Take 2 mL of peripheral blood and process it within 3 hours after obtaining the blood, including the following steps:
[0070] (1) 1600g, centrifuge peripheral blood for 10min, separate into blood cells and plasma (supernatant), and store blood cells at -80°C;
[0071] (2) The plasma sample was centrifuged for the second time at 16,000 g for 10 min, and the supernatant was transferred to a storage tube and stored at -80°C.
[0072] 2. In this example, nucleic acid extraction or purification (ZD-YL-Midi-40) was used for cfDNA extraction. The optimized extraction steps are as follows:
[0073] (1) Take a clean 10mL centrifuge tube, add 10μL ProteinaseK, then add 1mL plasma ...
Embodiment 2
[0086] Example 2 Detection of EGFR L858R and 19Del sites of ctDNA extracted from peripheral blood of patients with lung cancer
[0087] The droplet digital PCR system (droplet digital PCR) performs droplet treatment on samples before traditional PCR amplification, and divides the reaction system containing nucleic acid molecules into tens of thousands of nanoliter droplets, each of which may or may not Contain nucleic acid target molecules to be detected, or contain one to several nucleic acid target molecules to be detected. After PCR amplification, the fluorescence signal of each droplet is analyzed one by one. The droplet with fluorescent signal is interpreted as 1, and the droplet without fluorescent signal is interpreted as 0. According to the principle of Poisson distribution and the number of positive droplets The initial copy number or concentration of the target molecule can be obtained by ratio. Compared with the traditional PCR technology, the droplet digital PCR m...
Embodiment 3
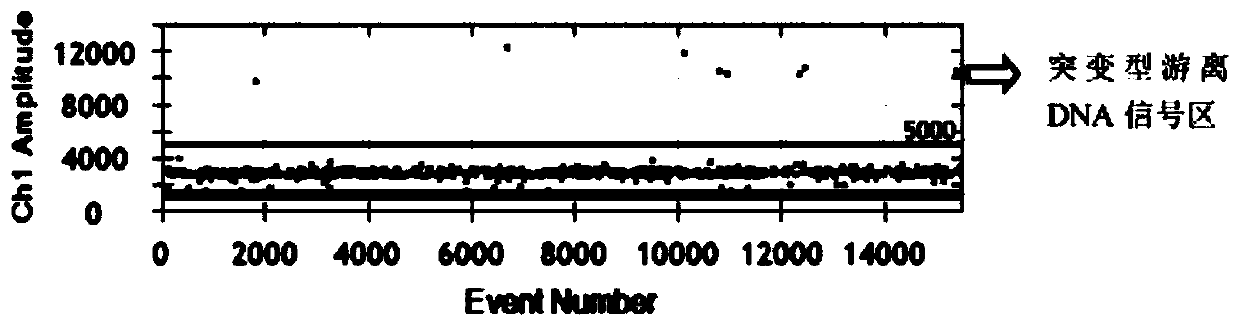
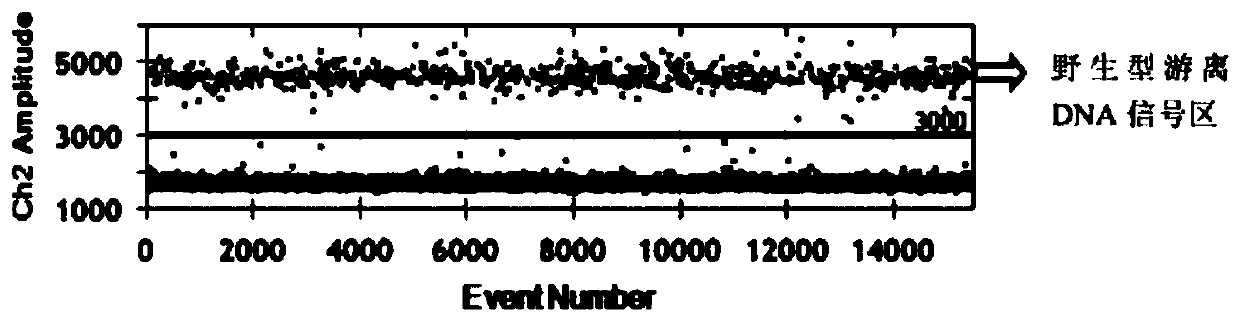
[0122] Example 3 Analysis of fluorescence signal values at EGFR L858R and EGFR 19Del sites
[0123] 1. Fluorescence signal analysis of EGFR L858R locus
[0124] (1) Open the QuantaSoft software, import the sample detection data, and select the Analyze module to start analyzing the data.
[0125] (2) First of all, it is necessary to confirm that the number of droplets generated by all sample types must be above 10,000, and only the CV value of the data detected within this range can be controlled within 1.6%. Then select 1D Amplitude to determine the fluorescence threshold of the mutant probe and the wild-type probe, and use the blank control group and the wild-type control group to delineate the threshold of the mutant probe and the wild-type probe. figure 2 , image 3 It shows that the EGFR L858R probe designed in the present invention has good specificity, and can clearly distinguish wild-type micro-droplets from mutant-type micro-droplets.
[0126] (3) The QuantaSoft ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com