Method for clustering nucleic acid sequences, equipment and storage medium
A nucleic acid sequence and sequence technology, applied in the field of computer equipment and computer-readable storage media, can solve problems such as incomplete information and affecting the results of species analysis, and achieve the effects of ensuring authenticity, reducing errors, and ensuring reliability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0115] This example constructs a specific implementation of the above technical solution, and uses simulated data to compare the results of this patent solution with Mothur and CD-HIT.
[0116]Among them, Mothur is a hierarchical clustering method. Its principle is to calculate the distance between two sequences, merge the two sequences with the closest distance into a cluster (cluster), and then form the cluster as a sequence, repeat the above Steps, until the distance between sequences or clusters is greater than the threshold and cannot be merged. In this embodiment, refer to the document Introducing mother: Open-Source, Platform-Independent, Community-Supported Software for Describing and Comparing Microbial Communities (Patrick D.Schlossetal. APPLIED AND ENVIRONMENTAL MICROBIOLOGY, Dec.2009, Vol.75, No.23 , p.7537-7541) to obtain the results of cluster analysis, as shown in Table 1.
[0117] CD-HIT is a heuristic clustering method. The basic method is to first take the ...
Embodiment approach
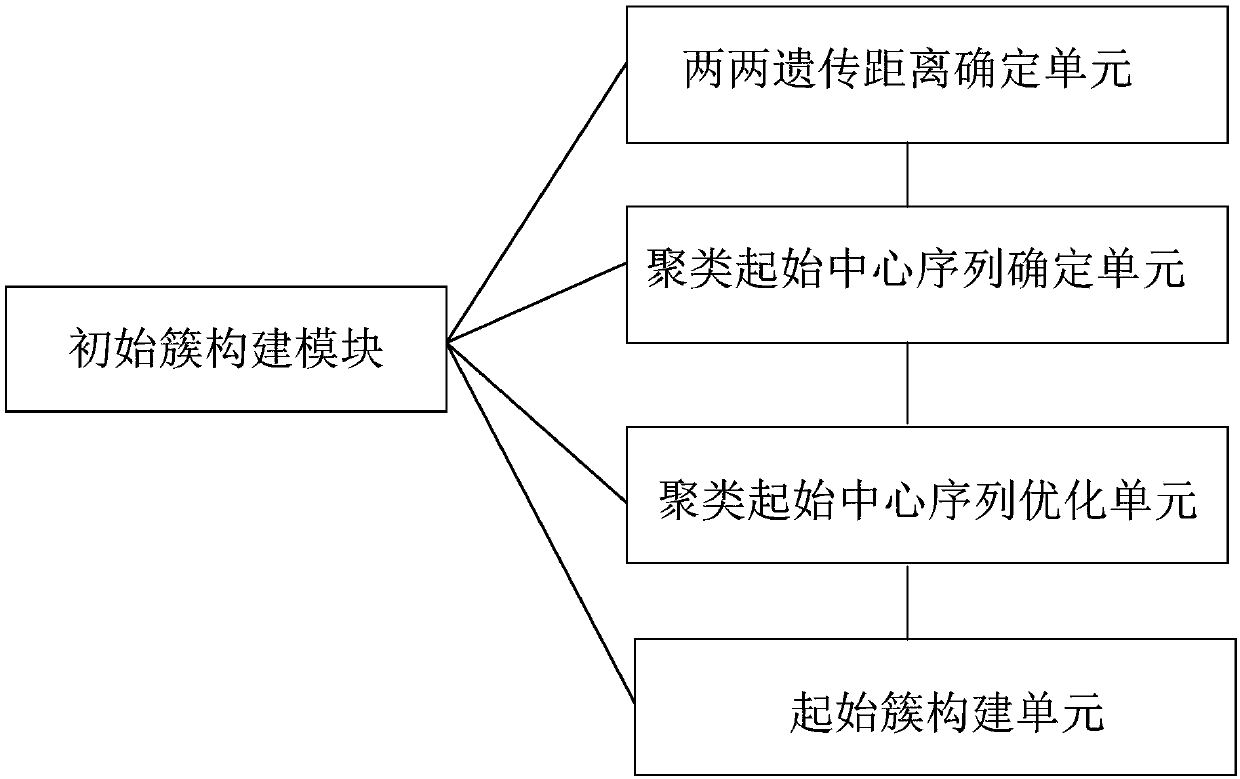
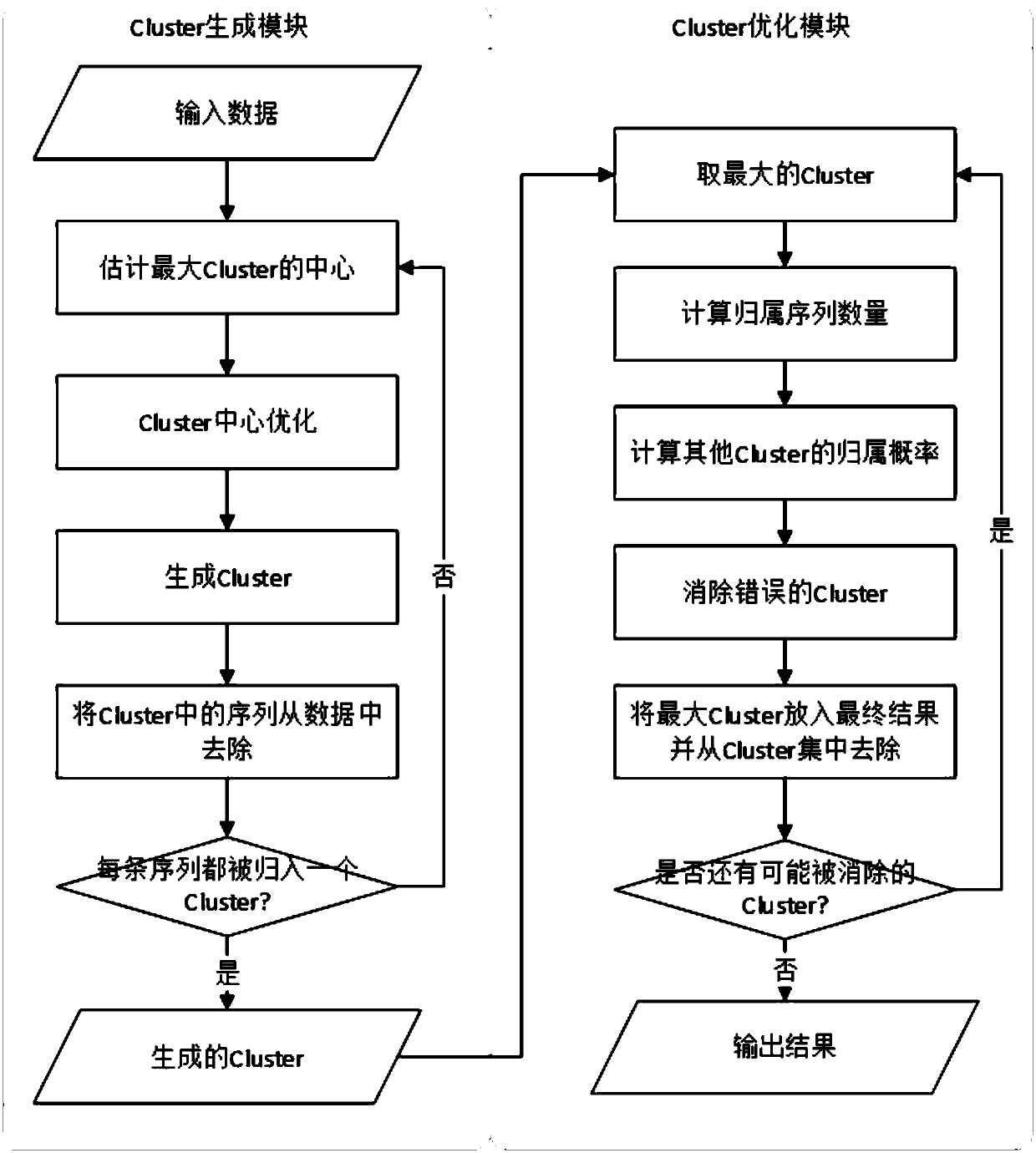
[0119] as attached image 3 shown. attached image 3 A flowchart for clustering multiple nucleic acid sequences is provided. It mainly includes cluster generation module and cluster optimization module. Among them, the cluster generation module includes the following processes:
[0120] First input the sequencing data, then estimate the largest cluster center, optimize the cluster center on this basis, and generate clusters. Then remove the sequences already contained in the cluster from the cluster, check whether each sequence is classified into a cluster, if not, re-estimate the largest cluster center and perform another cycle until each sequence is classified into the same cluster to generate different clusters.
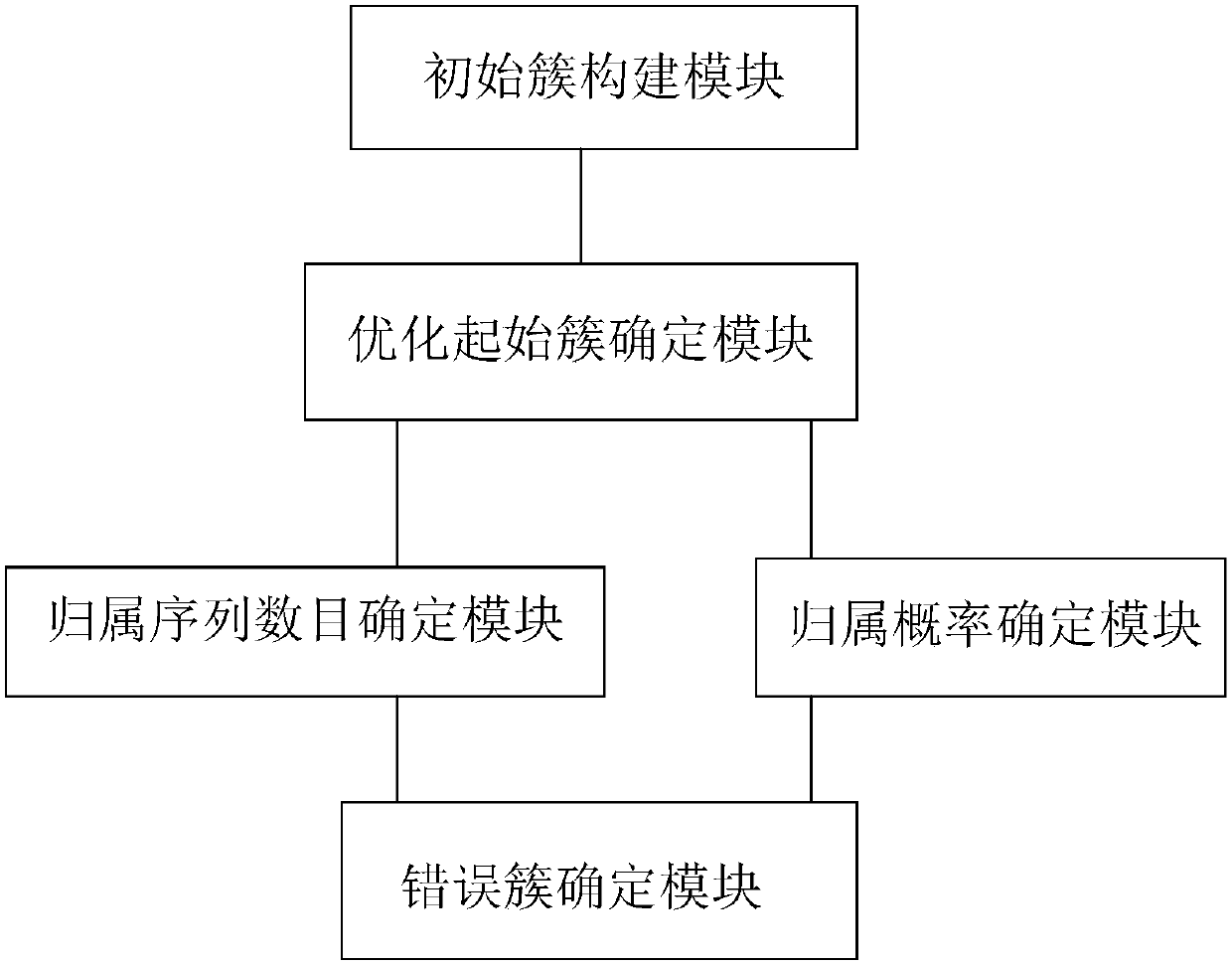
[0121] The cluster optimization module includes the following processes:
[0122] Take the largest cluster generated, calculate the number of belonging sequences and the belonging probabilities of other clusters, then eliminate the wrong cluster, and then re...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com