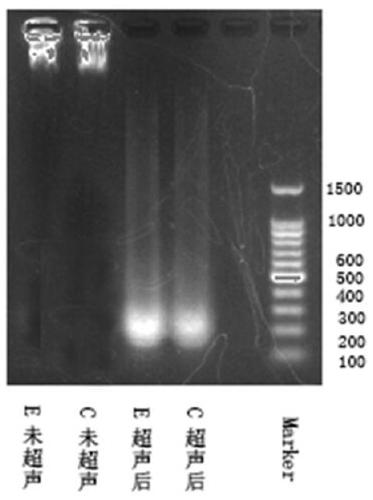
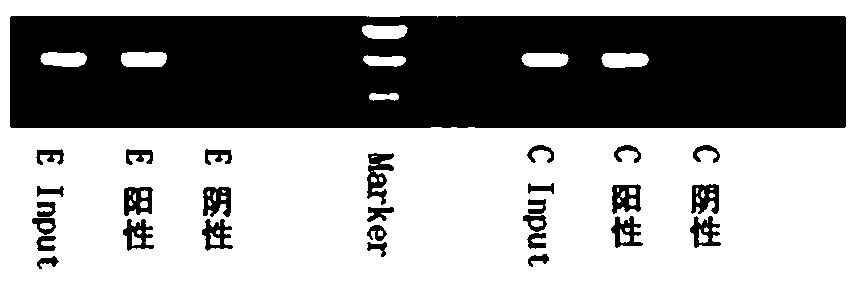
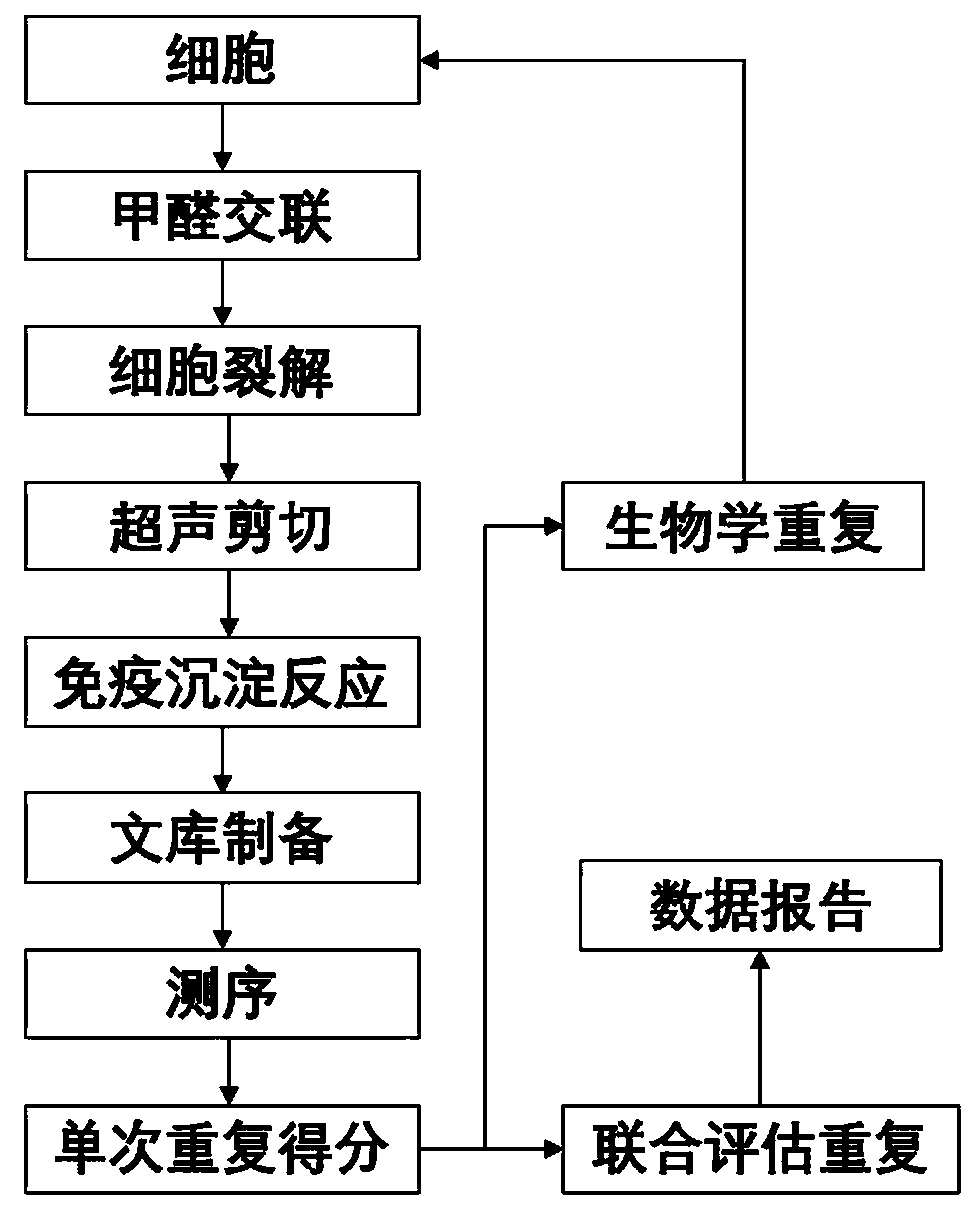
Identification method of BPDE adduct gene
An identification method and gene technology, applied in the field of identification of BPDE adducted genes, can solve the problems of high-throughput sequencing false positives, information loss, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
[0083] A kind of identification method of BPDE addition gene, comprises the following steps:
[0084] 1) Observe under the microscope that the degree of cell confluence in the culture medium grows to 70%-80%, and the cells are in good condition at this time.
[0085] In this step, confluency is a measure of the density of cells in the dish. This indicator is a well-known concept in the art. Only when the cell confluence reaches 70%-80%, it means that the cell is in good condition, then proceed to the following steps.
[0086] 2) Add formaldehyde to the medium to fix the cells in the formaldehyde, vortex the culture dish to mix the formaldehyde in the medium, and let stand at room temperature for 10 minutes.
[0087] In this example, the preferred method of adding formaldehyde is to add methanol solution (the solvent is deionized water), and the preferred concentration of formaldehyde solution is: add 275 μL of 37wt% formaldehyde solution per 10 mL of medium. Try to avoid di...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com