Library establishment method for metagenome sequencing, kit and sequencing method
A metagenomics and kit technology, applied in the field of sequencing, can solve serious problems in identification, increase the number of PCR cycles, aggravate amplification bias, etc., reduce the initial amount of library construction, reduce PCR amplification bias, The effect of improving the success rate of database construction
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0044] Example 1 The official library construction method of SQK-RPB004 kit
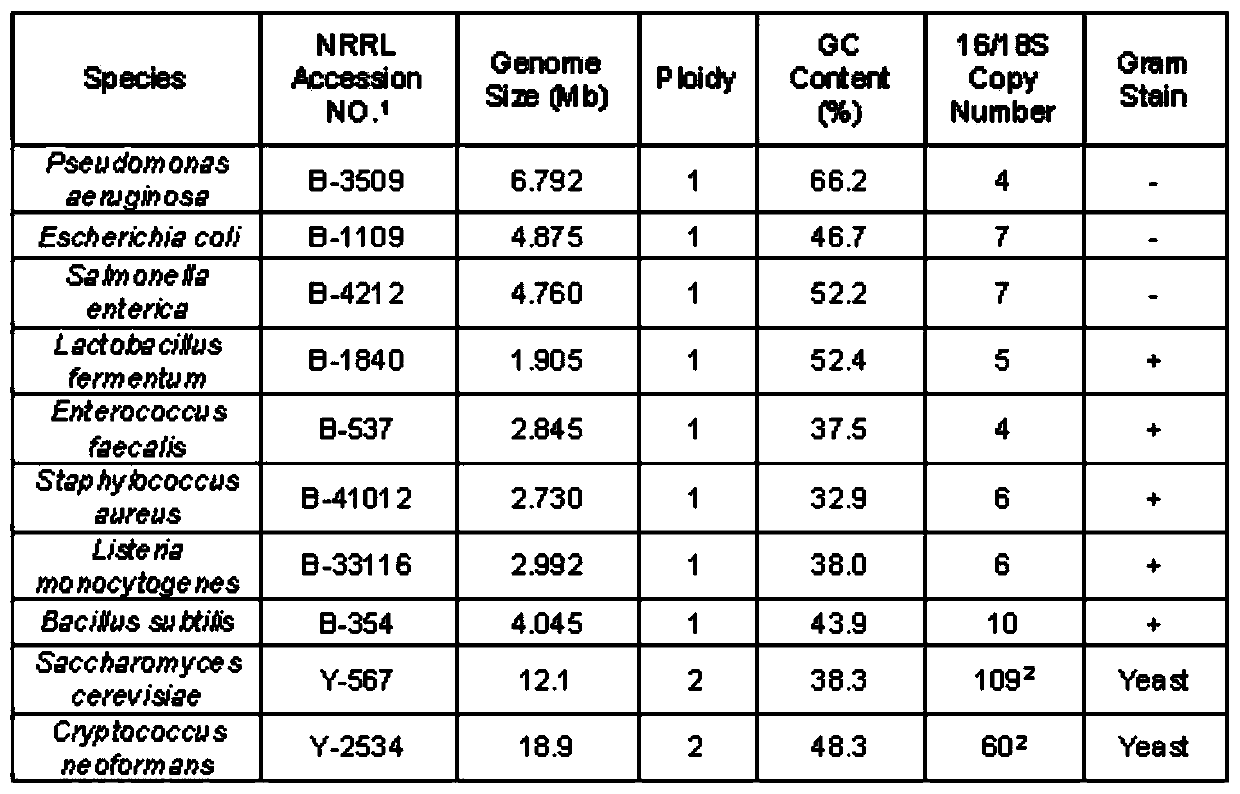
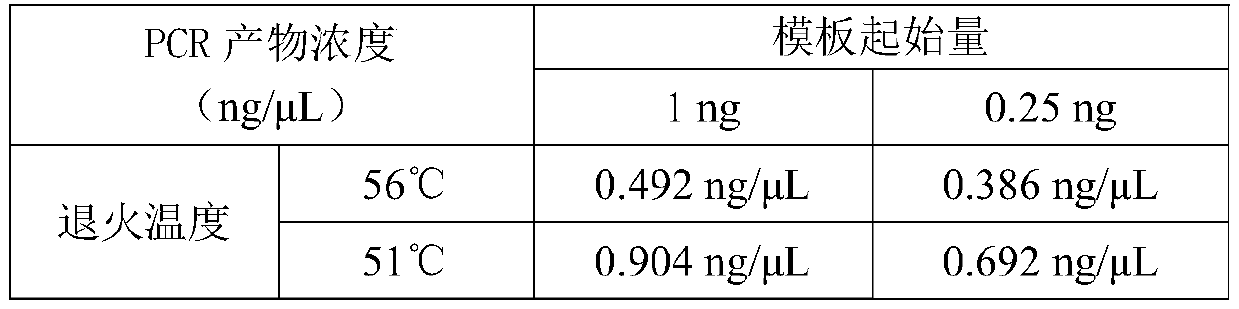
[0045] Take the zymo standard as the sample and build the library according to the official process provided by the SQK-RPB004 kit. The initial amount of sample DNA is 1ng and 0.25ng (3μL) respectively, the FRM fragmentation mixture is 1μL, and the RLB rapid barcode primer is 1μL , The annealing temperature was 56°C and 51°C (see Table 1). After 14 cycles are completed according to the official process, the nucleic acid concentration of the PCR product is less than 1ng / μL (Table 2), which is far from meeting the requirements of the computer (according to the official instructions, the total nucleic acid amount at the mixing stage needs to be at least 100ng, and considering the purification loss , The mixing amount of a single sample should not be less than 200ng in the mixed library stage, that is, calculated with a 50μL system, the concentration should not be less than 4ng / μL).
[0046] Table 1. SQK-RPB...
Embodiment 2
[0050] Example 2 Grady library construction method
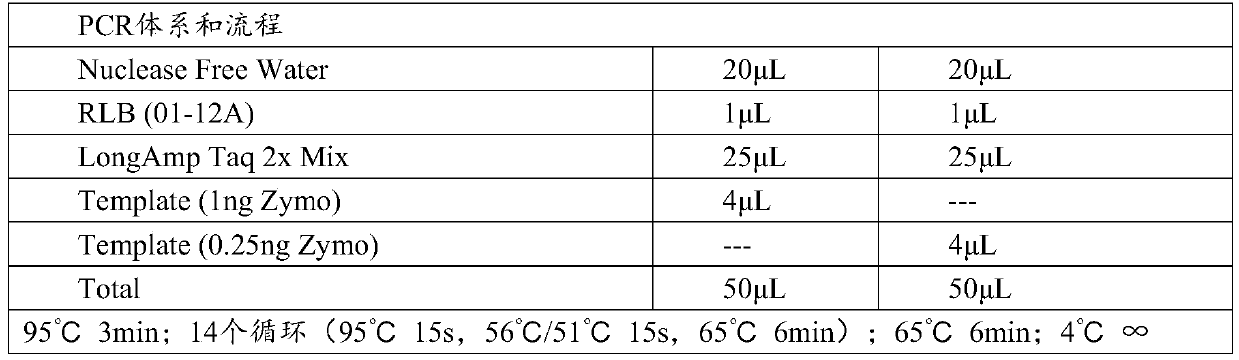
[0051] 1. Build the library according to the Grady method, in which 7.5μL DNA template and 2.5μL FRM fragmentation mixture are mixed, the number of cycles is increased to 25 cycles, and the extension time is shortened to 4mins. Still taking 1ng and 0.25ng DNA template as the starting amount, the RLB fast barcode primer dosage is 1μL, try 56°C and 51°C respectively (see Table 3 for specific PCR conditions). The result of library construction shows that after 25 cycles, the amount of nucleic acid of PCR product meets the computer requirements (see Table 4).
[0052] Table 3. SQK-RPB004Grady process of Zymo standard product tried PCR conditions
[0053]
[0054] Table 4. Results of 25-cycle PCR products of the SQK-RPB004Grady process of Zymo standard products
[0055]
[0056] 2. Follow the official process to mix the library on the computer and perform bioinformatics analysis according to the standard process. However, the sequencin...
Embodiment 3
[0063] Example 3 Optimization of library construction system (RLB primers and PCR reaction cycles)
[0064] On the basis of Example 2, the PCR system was optimized for RLB rapid barcode primers and the number of amplification cycles to improve the success rate of library construction: Increase the amount of RLB primers incorporated to 2 μL, and reduce the number of PCR amplification cycles to 14 And 20, check whether the database can be built (see Table 7 for specific PCR conditions). The results show that when the number of cycles is 14, the amount of nucleic acid of the PCR product cannot be used on the machine (see Table 8). When the number of cycles is 20, the amount of nucleic acid in the PCR product meets the computer requirements (see Table 9). Therefore, comparing the results of the 1μL dosage of RLB fast barcode primer, 2μL RLB primer (20 cycles) is the optimized amplification condition.
[0065] Table 7. SQK-RPB004Grady process improvement of Zymo standard products try ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com