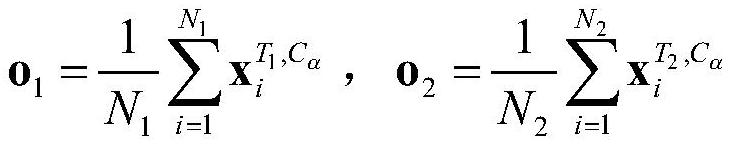A Differential Evolution-Based Method for Predicting the Structure of Protein Dimers
A prediction method, differential evolution technology, applied in proteomics, genomics, instrumentation, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
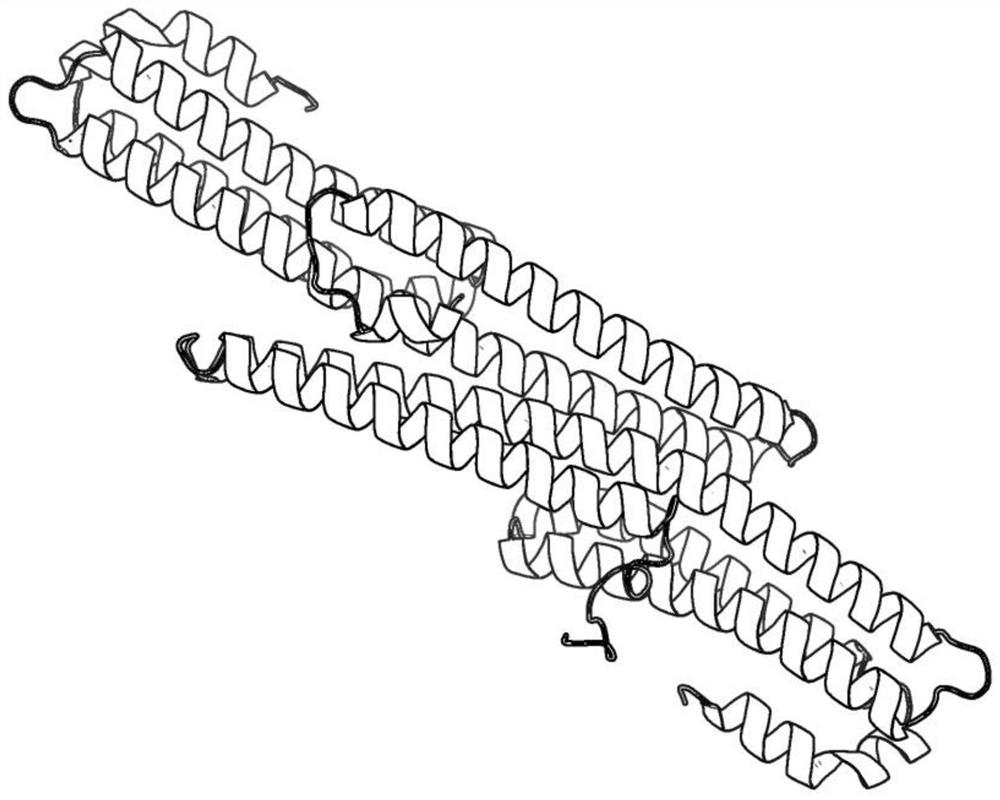
[0039] The present invention will be further described below in conjunction with the accompanying drawings.
[0040] refer to figure 1 and figure 2 , a protein dimer structure prediction method based on differential evolution, comprising the following steps:
[0041] 1) Input the sequence information of the two chains in the protein dimer to be predicted, denoted as Chain respectively 1 with Chain 2 ;
[0042] 2) For the input sequence information Chain 1 with Chain 2 , using the I-TASSER server (http: / / zhanglab.ccmb.med.umich.edu / I-TASSER / ) to predict the corresponding three-dimensional spatial structure information, denoted as T 1 with T 2 ;
[0043] 3) Calculate T 1 with T 2 The coordinates of the center point of , denoted as o 1 with o 2 :
[0044]
[0045] Among them, N 1 with N 2 Respectively represent T 1 with T 2 The number of amino acids contained in and Respectively represent T 1 with T 2 The central carbon atom C of the i-th amino acid in ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com