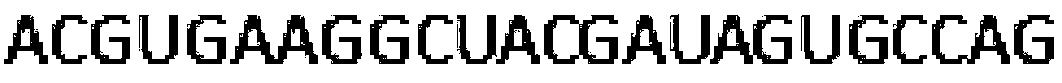Simulated annealing-based nucleic acid structure prediction method
A technology of simulated annealing and prediction methods, applied in the field of bioinformatics engineering, which can solve problems such as high time complexity and space complexity, and ineffective RNA sequences
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0060] In order to make the present invention easier to understand, the specific embodiments of the present invention will be further described below.
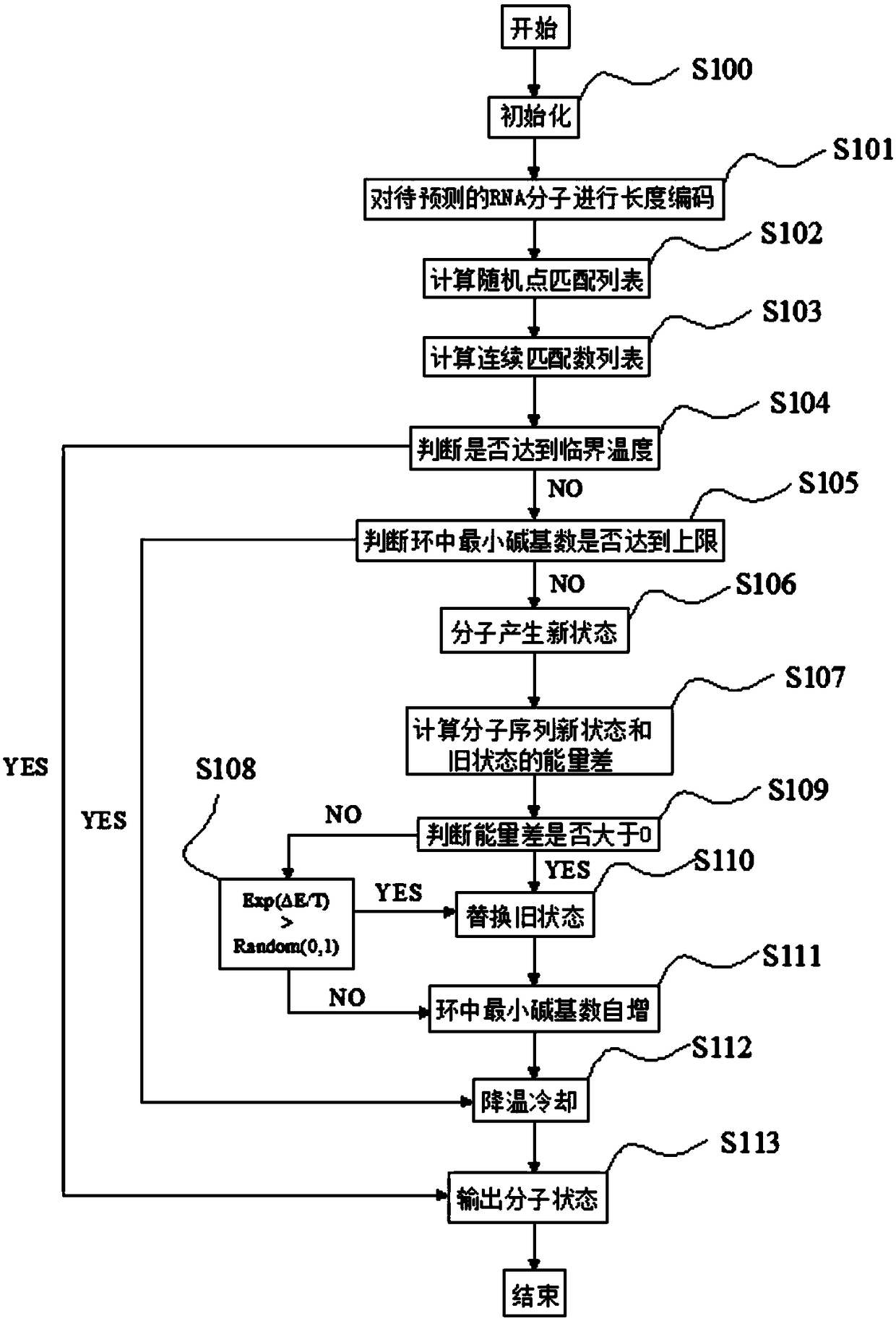
[0061] Such as image 3 Shown, a nucleic acid structure prediction method based on simulated annealing, comprising the following steps:
[0062] S100, set the minimum number of stem regions, the minimum number of bases in the ring, and the initial temperature value T 0 , critical temperature T 临 , molecular new state, molecular old state, the maximum number of pseudoknot cross-grouping; the minimum number of stem regions is set to 2, since the RNA sequence cannot be folded violently, there must be at least three bases in the loop, so the minimum number of bases in the loop needs to be The default setting is 3, the initial temperature value T 0 Can be set to 1500.
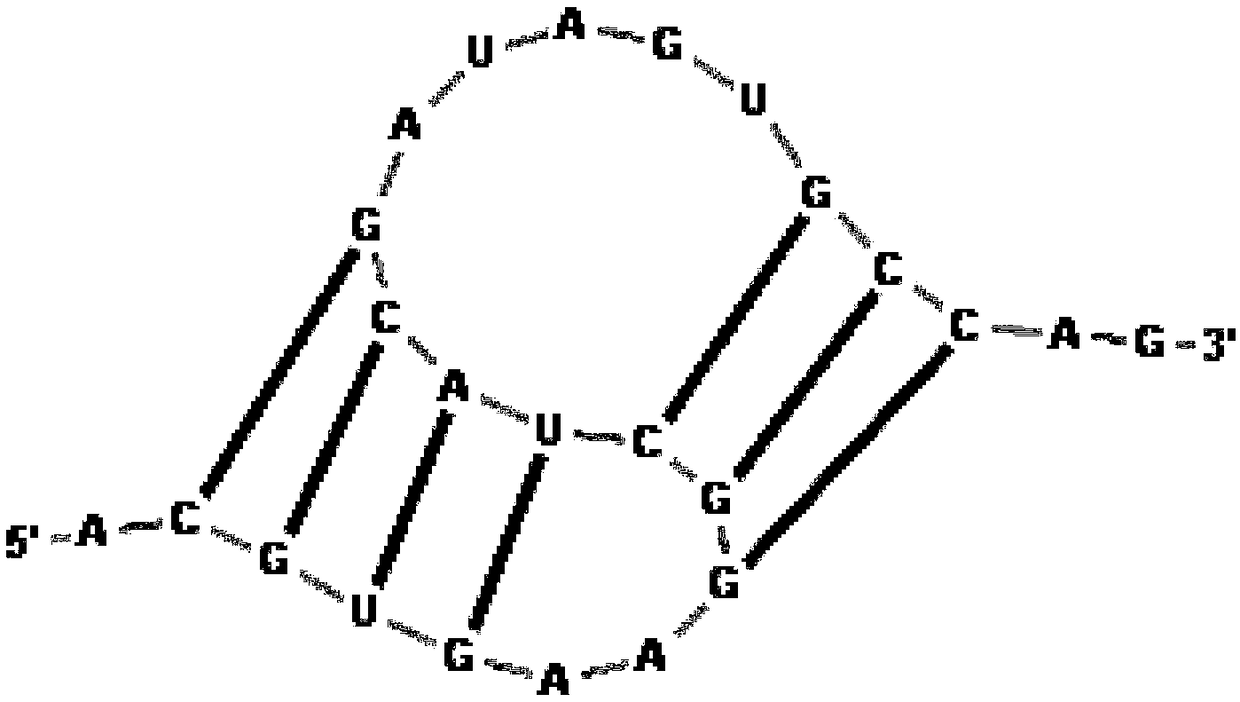
[0063] S101, denote the RNA sequence S as x 1 x 2 x 3 … x n , where x i ∈{A,C,G,U}, the length of the sequence is n, 1≤i≤j≤n; the length of the RNA sequenc...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com