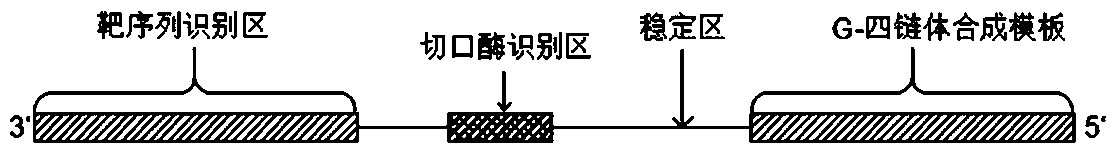
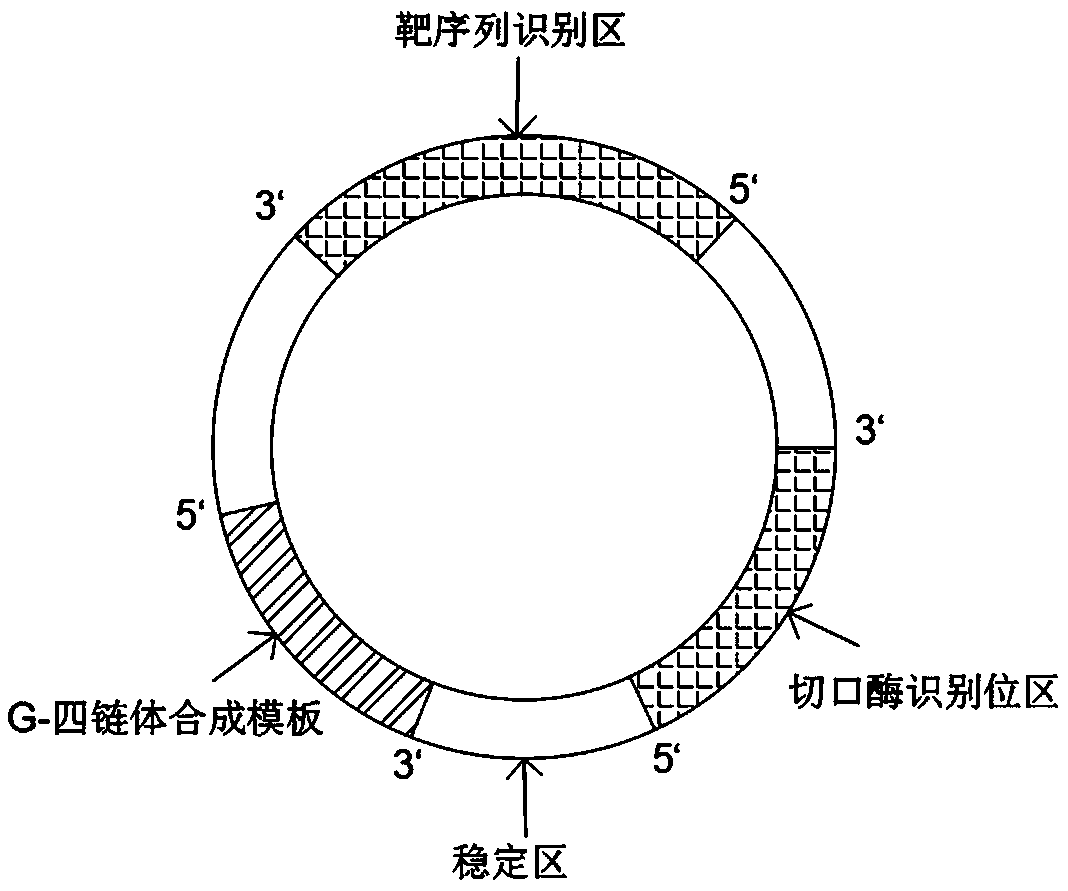
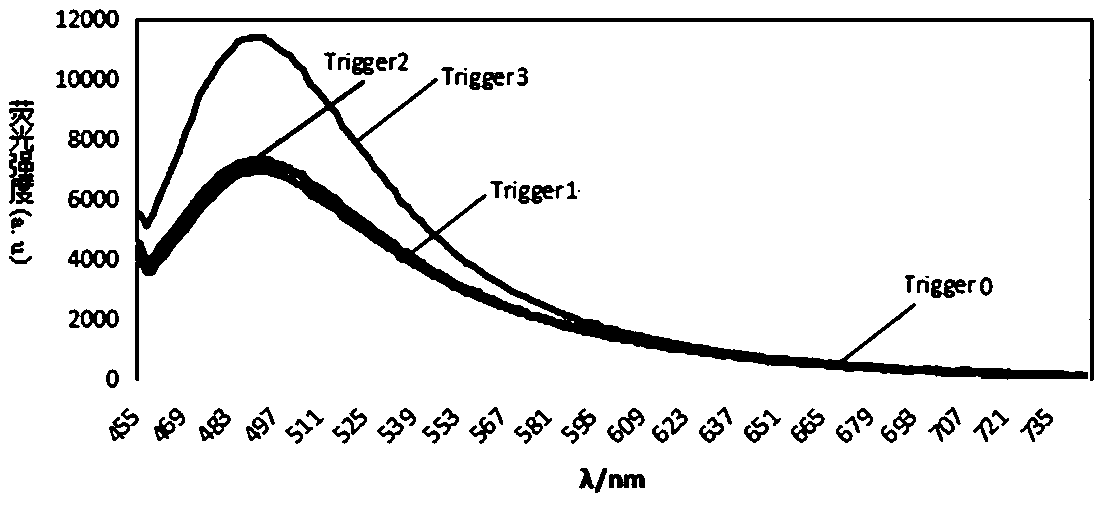
DNA molecular machine and nucleic acid detection method
A DNA molecule and detection method technology, applied in biochemical equipment and methods, microbiological determination/inspection, etc., can solve the problems of cumbersome operation, complex sample components, and low target DNA content, so as to reduce processing steps and broaden the scope of application , the effect of high sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0080] Step 1: Synthesize the DNA molecular machine and configure it into a solution with a concentration of 20nM,
[0081] Step 2: Pulverize mouse liver cancer tissue at room temperature, filter and extract the juice, and prepare a sample solution with a concentration of 2 μM,
[0082] Step 3: Take 4 μL of sample solution and 2 μL of DNA molecular machine solution, mix evenly by micro-oscillation to obtain a mixed solution,
[0083] Step 4: Add 10 μL buffer, 6 μL polymerase, 6 μL nickase, 6 μL Thioflavin T, 6 μL dNTP, 6 μL Mg 2+ Add to the mixed solution obtained in step 3 to obtain a reaction solution.
[0084] Wherein, the above-mentioned dNTP concentration is 0.12mM; the polymerase concentration is 0.112U / uL; the nickase concentration is 0.01U / uL; the thioflavin T concentration is 0.1uM; HCL, tween20 at a concentration of 0.1%, KCl at a concentration of 130 mM, (NH4) at a concentration of 10 mM 2 SO 4 and NaCl at a concentration of 30 mM; Mg 2+ The concentration is 1m...
Embodiment 2
[0101] Step 1: Synthesize the DNA molecular machine and configure it into a solution with a concentration of 60nM,
[0102] Step 2: Grinding the mouse liver cancer tissue at room temperature, filtering and extracting the juice, and preparing a sample solution with a concentration of 8 μM,
[0103] Step 3: Take 8 μL of sample solution and 4 μL of DNA molecular machine solution, mix evenly by micro-oscillation to obtain a mixed solution,
[0104] Step 4: Add 40 μL buffer, 10 μL polymerase, 10 μL nickase, 10 μL Thioflavin T, 10 μL dNTP, 10 μL Mg 2+ Add to the mixed solution obtained in step 3 to obtain a reaction solution.
[0105] Wherein, the above-mentioned dNTP concentration is 0.50mM; the polymerase concentration is 0.236U / uL; the nickase concentration is 0.06U / uL; the buffer solution includes Tris HCL with a concentration of 70mM, pH9.0, and tween20 with a concentration of 0.6%. KCl at a concentration of 160mM, (NH4) at a concentration of 60mM 2 SO 4 , NaCl at a concent...
Embodiment 3
[0115] Step 1: Synthesize the DNA molecular machine and configure it into a solution with a concentration of 100nM,
[0116] Step 2: Pulverize mouse liver cancer tissue at room temperature, filter and extract the juice, and prepare a sample solution with a concentration of 2 μM,
[0117] Step 3: Take 4 μL of the sample solution and 4 μL of the DNA molecular machine solution, and mix evenly by micro-oscillation to obtain a mixed solution.
[0118] Step 4: Add 20 μL buffer, 6 μL polymerase, 7 μL nickase, 10 μL Thioflavin T, 3 μL dNTP, 36 μL Mg 2+ Add to the mixed solution obtained in step 3 to obtain a reaction solution.
[0119] Wherein, the above-mentioned dNTP concentration is 0.3mM; the polymerase concentration is 0.212U / uL; the nickase concentration is 0.03U / uL; the thioflavin T concentration is 0.1uM; HCL, tween20 at a concentration of 0.4%, KCl at a concentration of 145mM, (NH4) at a concentration of 45mM 2 SO 4 , a concentration of 46mM NaCl; Mg 2+ The concentration...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com