Monitoring and diagnosis for immunotherapy, and design for therapeutic agent
A device, sequencing technology, applied in the determination/inspection of microorganisms, for the analysis of 2D or 3D molecular structure, bioreactors/fermenters for specific purposes, etc., can solve the problem of cancer that has not been fully studied targeting mutant gene products Immunotherapy and other issues
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0197] (Example 1: Sorting of mutant peptides and detection of immunogenicity - against tumors in human subjects)
[0198] (analyze)
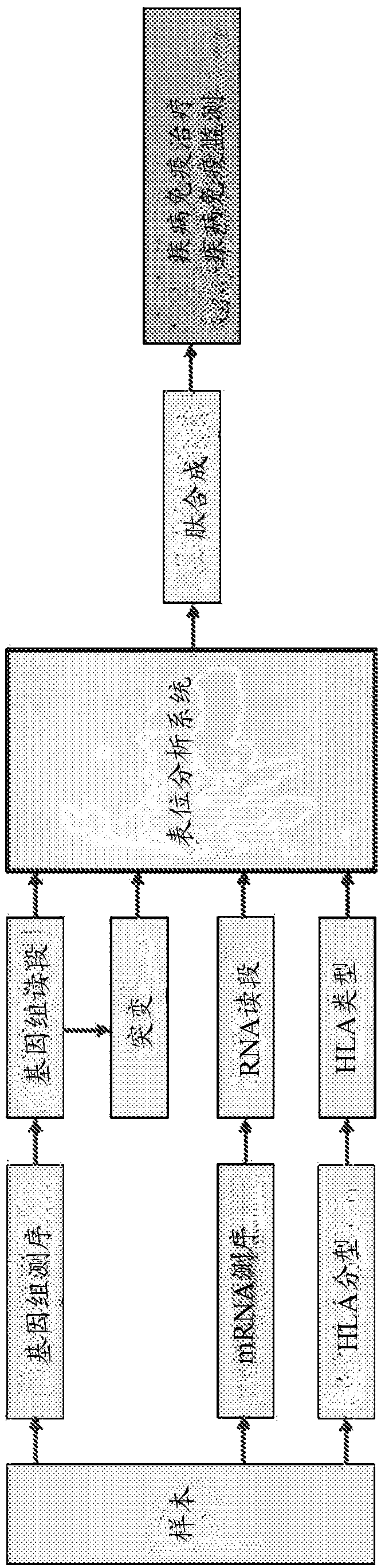
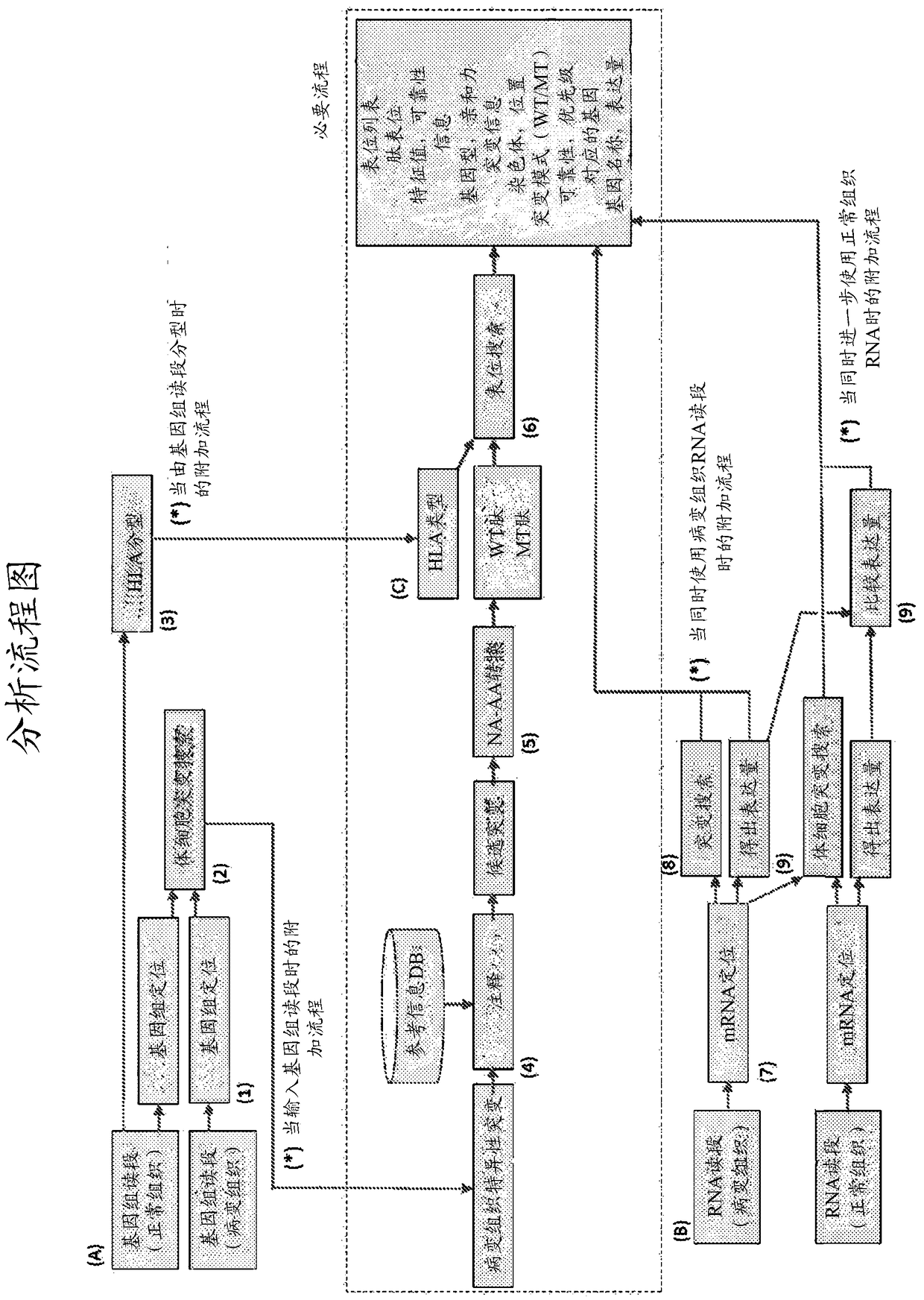
[0199] based on figure 2 Mutant peptides were sorted using the protocol exemplified in .
[0200] Specifically, do the following.
[0201] (1) Subjects of the following groups were used.
[0202] Target patient: 65-year-old Japanese male lung cancer patient
[0203] Prior to performing the Examples, the following HLA types were identified by typing using the Luminex assay.
[0204] HLA type: HLA-A*02:01,24:02
[0205] (2) Extract DNA from normal tissue and tumor tissue. Exome sequencing was performed with the HiSeq 2000 sequencer from Illumina using the TruSeq PE kit. The equipment used was a sequencer HiSeq 2000 from Illumina, and the software used was the control software of the same sequencer.
[0206] (Exome Read Mapping)
[0207] Next, bwa was used with the following parameters to map exome reads from normal and tumor tissues res...
Embodiment 2
[0318] (Example 2: Sorting of mutant peptides and detection of immunogenicity - for mice)
[0319] In this example, the same experiment can be performed when using mice.
[0320] Its procedure is as follows.
[0321] 1. Neoantigens can be searched for spontaneously, chemically and radiation-induced tumors (cancer, sarcoma or leukemia) in all syngenic mouse strains. In this example, a mouse strain was used: C57BL / 6 (MHC haplotype H2 b ). Tumors: B16 melanoma cells were used as tumors.
[0322] 2. Collect tissue from the cancer site of the cancer-bearing mouse, and collect the same organ / tissue as the tumor site in the normal mouse, wherein the tumor site and the non-tumor site (for example, tumor of colon cancer) are collected in the cancer-bearing mouse sites and non-tumor sites). Normal tissues were collected from normal mice when the mice were from the same strain.
[0323] 3. Extract DNA and RNA from collected tissues / organs for exome-seq and RNA-seq analysis.
[032...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com