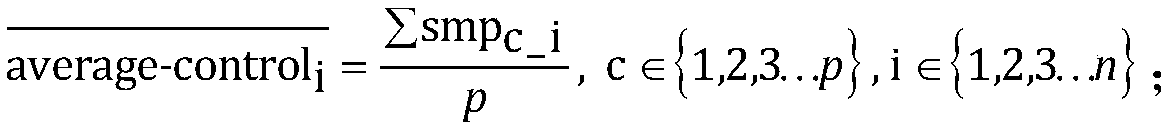Single exon copy number variation predicting method based on target area sequencing
A technology of copy number variation and target region, which is applied in the field of single exon copy number variation prediction, can solve problems such as complex analysis process, and achieve the effect of simple method, low cost, and reduced analysis complexity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
specific Embodiment 1
[0099] Three positive samples known to have exon-level copy number variation were analyzed, and the exon copy number variation information of the three positive samples was as follows.
[0100]
[0101]
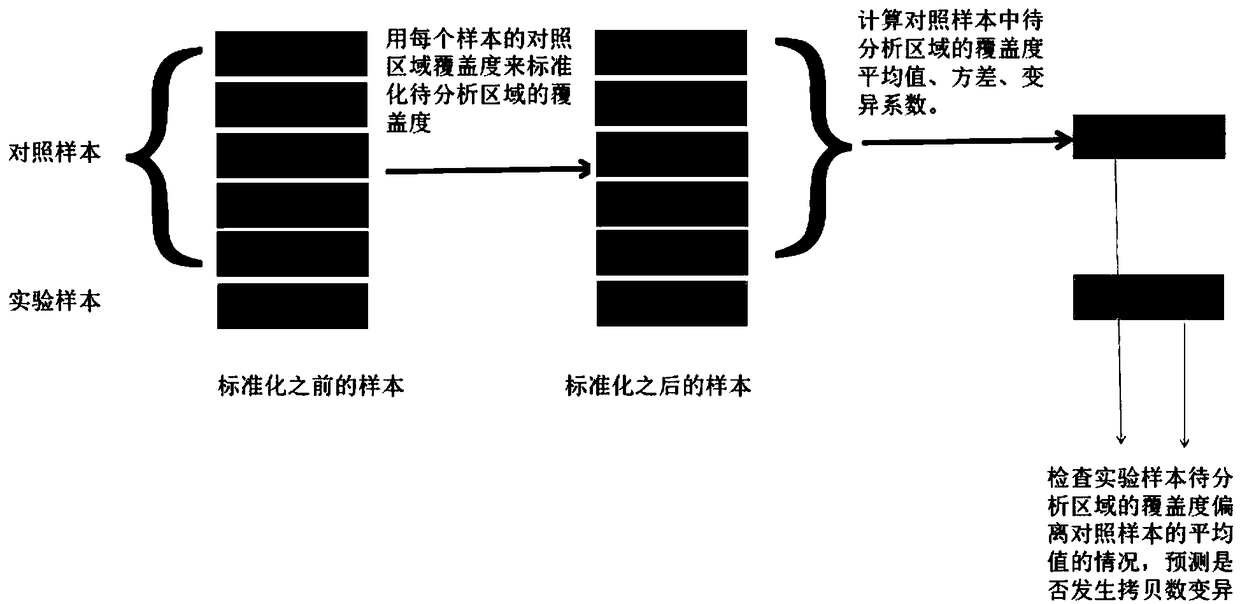
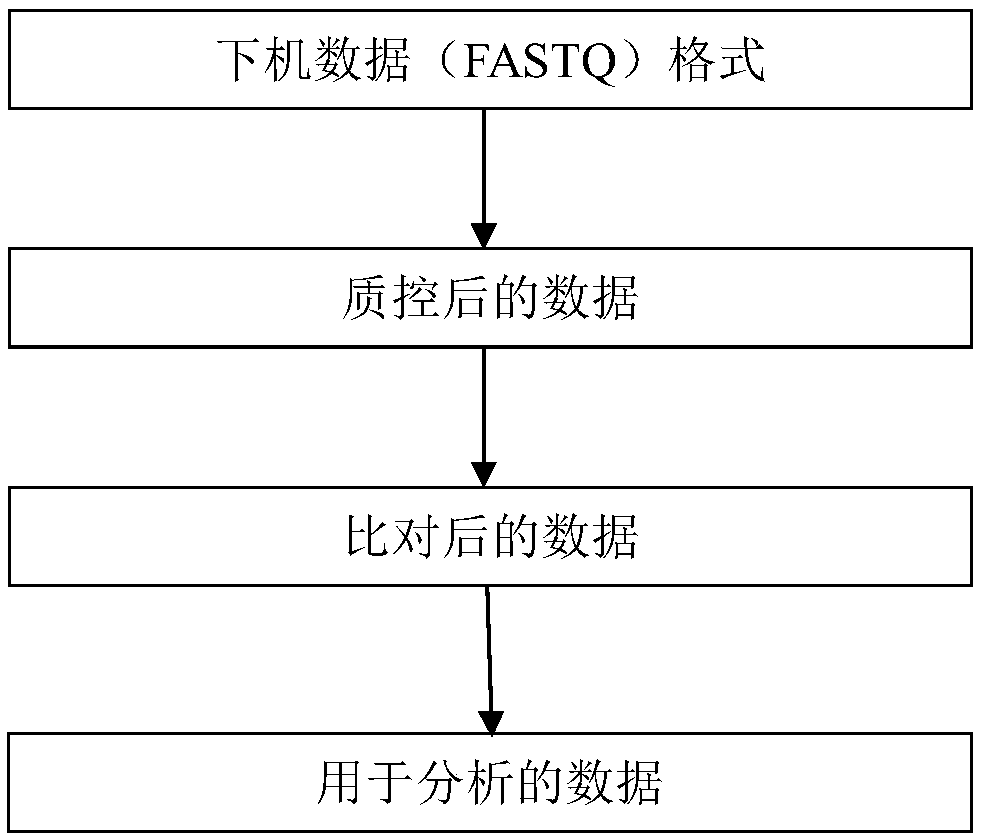
[0102] Three positive samples and five negative control samples were subjected to exome sequencing to obtain sequencing data. Perform quality control on the sequencing data, compare it to the hg19 reference genome, and use picard to deduplicate and sort the aligned reads. The software used is trommomatic, bwa, picard. The statistical information of each sample is as follows:
[0103] sample
[0104] Use the software bedtools to count the coverage of each exon, and then normalize the coverage of the exons to be analyzed for each sample. The coverage information of the five control samples was formed into a control sample group, and the positive samples were analyzed one by one. The test results are as follows:
[0105] sample
[0106] The overall ...
specific Embodiment 2
[0112] For the prediction of deletion variants, we also achieved good results, because the sequencing coverage of the exons in the samples with deletions was almost zero. Taking the NA05169 sample as an example, 40 exons in this sample have copy number deletion mutations, and the predicted results are as follows:
[0113]
[0114]
[0115] This method detected all 40 deletion variants in the NA05169 sample.
[0116] In summary, this method does not use GC content to correct the prediction of copy number duplication and deletion variation, nor does it perform complex modeling like other software for prediction. Only 5 control samples are used, that is, the duplication and deletion Missing mutations were predicted, showing the good application performance of the algorithm. The source of the data is the data obtained from the sequencing of the existing target region, and no additional experimental cost is required.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com